Search Count: 25
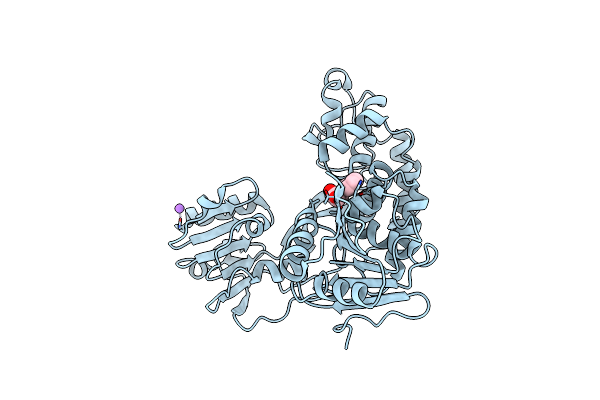 |
X-Ray Crystal Structure Of Ns3 Helicase From Hcv With A Bound Fragment Inhibitor At 2.08 A Resolution
Organism: Hepatitis c virus
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2015-12-23 Classification: HYDROLASE/HYDROLASE INHIBITOR |
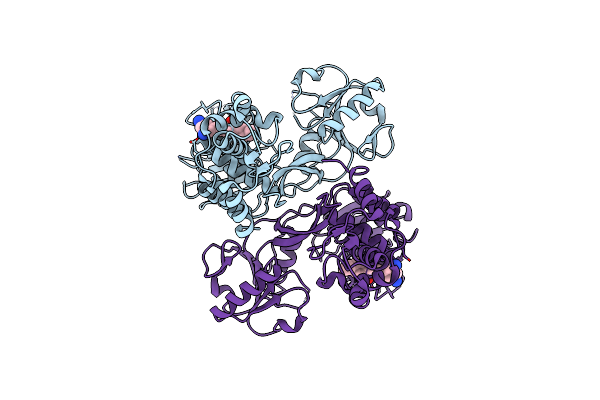 |
X-Ray Crystal Structure Of Ns3 Helicase From Hcv With A Bound Inhibitor At 2.42 A Resolution
Organism: Hepatitis c virus
Method: X-RAY DIFFRACTION Resolution:2.42 Å Release Date: 2015-12-23 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: 3VZ |
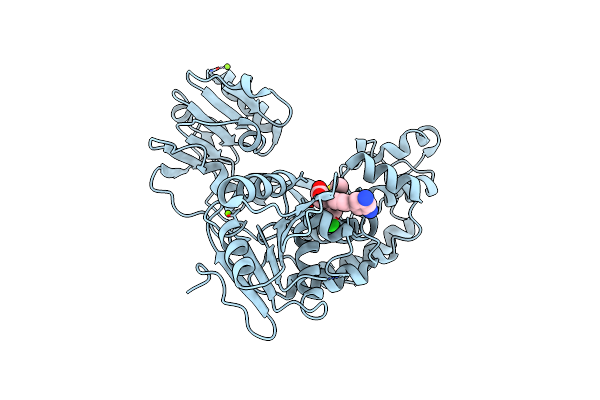 |
Organism: Hepatitis c virus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2014-09-24 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: I79, MG, CL |
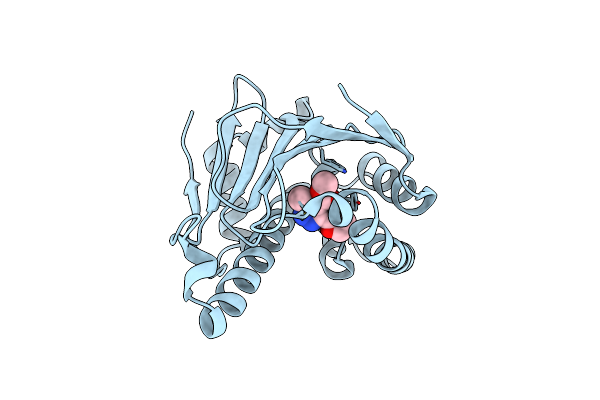 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2010-07-14 Classification: CHAPERONE Ligands: 37D |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2010-07-14 Classification: CHAPERONE Ligands: 42C |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2010-07-14 Classification: CHAPERONE Ligands: 42C, 37D |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2010-07-14 Classification: CHAPERONE Ligands: Z64 |
 |
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2010-01-26 Classification: ISOMERASE Ligands: MN, PLP |
 |
X-Ray Crystal Structure Of Human Serine Racemase In Complex With Malonate A Potent Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2010-01-26 Classification: ISOMERASE Ligands: PLP, MN, MLI |
 |
X-Ray Crystal Structure Of Rat Serine Racemase In Complex With Malonate A Potent Inhibitor
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2010-01-26 Classification: ISOMERASE Ligands: PLP, MN, MLI |
 |
The Structure Of Mammalian Serine Racemase: Evidence For Conformational Changes Upon Inhibitor Binding
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2010-01-26 Classification: ISOMERASE Ligands: MLI, MN |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2009-08-04 Classification: OXIDOREDUCTASE Ligands: NAP, ACT, EDO |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2009-08-04 Classification: OXIDOREDUCTASE Ligands: NAP, ACT |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2009-06-30 Classification: CHAPERONE Ligands: MOJ |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2009-05-05 Classification: CHAPERONE Ligands: MO8 |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2007-12-11 Classification: HYDROLASE Ligands: MN, SO4, ACT, EDO, GOL |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2007-12-11 Classification: HYDROLASE Ligands: CO, PO4, ACT, EDO |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2007-12-11 Classification: HYDROLASE Ligands: MG, ATP |
 |
Structure Of Transhydrogenase (Di.R127A.Nad+)2(Diii.Nadp+)1 Asymmetric Complex
Organism: Rhodospirillum rubrum
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2006-02-28 Classification: OXIDOREDUCTASE Ligands: NAD, NAP |
 |
Structure Of Transhydrogenase (Di.S138A.Nadh)2(Diii.Nadph)1 Asymmetric Complex
Organism: Rhodospirillum rubrum
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2006-02-28 Classification: OXIDOREDUCTASE Ligands: NAI, NDP |

