Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
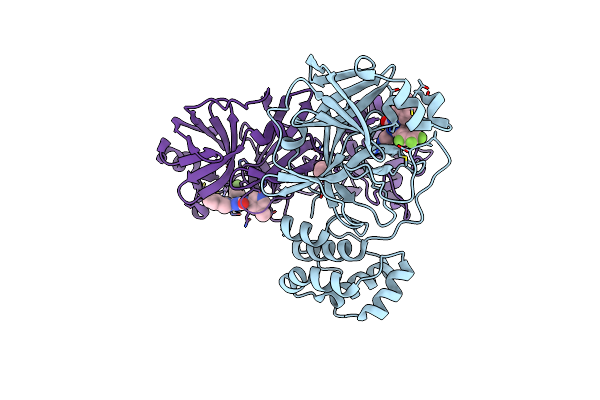 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: VIRAL PROTEIN Ligands: A1L7P, EDO |
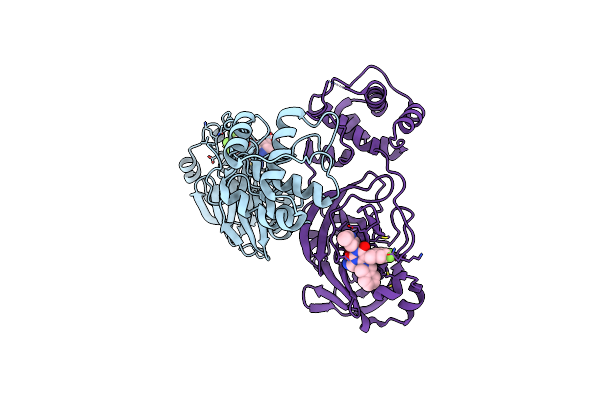 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: VIRAL PROTEIN Ligands: A1L7Q |
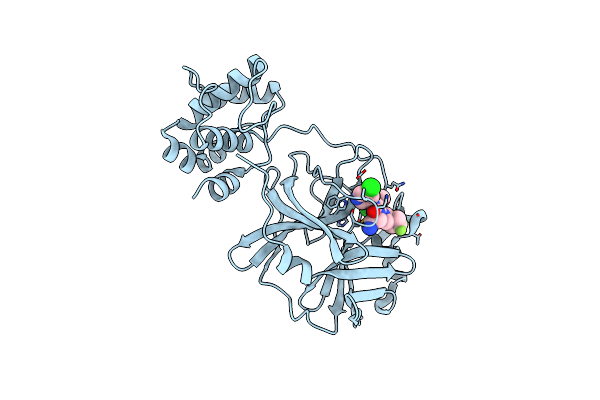 |
Crystal Structure Of Sars-Cov-2 3Cl Protease In Complex With Compound 17 (S-892216)
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: VIRAL PROTEIN Ligands: A1L7R |
 |
Cryo-Em Structure Of Calcium Sensing Receptor In Complex Gamma-Glutamyl-Valyl-Glycine As A Kokumi Substance
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-02-19 Classification: MEMBRANE PROTEIN |
 |
The Crystal Structure Of Sars-Cov-2 3Cl Protease In Complex With Compound 1
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-04-06 Classification: VIRAL PROTEIN/INHIBITOR Ligands: 7XB |
 |
The Crystal Structure Of Sars-Cov-2 3Cl Protease In Complex With Compound 3
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2022-04-06 Classification: Viral protein/inhibitor Ligands: 7YY |
 |
Organism: Sphingomonas sp
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2020-02-19 Classification: SUGAR BINDING PROTEIN |
 |
Organism: Sphingomonas sp. a1
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2020-01-29 Classification: TRANSPORT PROTEIN Ligands: EDO, CIT |
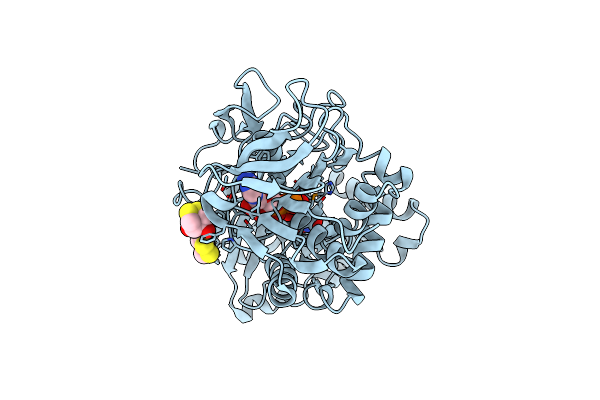 |
Crystal Structure Of The Modified Fructosyl Peptide Oxidase From Aspergillus Nidulans, Seleno-Methionine Derivative
Organism: Aspergillus nidulans
Method: X-RAY DIFFRACTION Resolution:2.61 Å Release Date: 2019-05-15 Classification: OXIDOREDUCTASE Ligands: FAD, D1D, SO4 |
 |
Crystal Structure Of The Modified Fructosyl Peptide Oxidase From Aspergillus Nidulans In Complex With Fsa, Seleno-Methionine Derivative
Organism: Aspergillus nidulans
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2019-05-15 Classification: OXIDOREDUCTASE Ligands: FAD, FSA, D1D, SO4 |
 |
Crystal Structure Of The Modified Fructosyl Peptide Oxidase From Aspergillus Nidulans With R61G Mutation
Organism: Aspergillus nidulans
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2019-05-15 Classification: OXIDOREDUCTASE Ligands: FAD, SO4, D1D |
 |
Crystal Structure Of The Modified Fructosyl Peptide Oxidase From Aspergillus Nidulans With R61G Mutation, In Complex With Fsa
Organism: Aspergillus nidulans
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2019-05-15 Classification: OXIDOREDUCTASE Ligands: FAD, FSA, D1D, SO4 |
 |
Crystal Structure Of The Modified Fructosyl Peptide Oxidase From Aspergillus Nidulans With 7 Additional Mutations, In Complex With Fsa
Organism: Aspergillus nidulans
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2019-05-15 Classification: OXIDOREDUCTASE Ligands: FAD, FSA |
 |
Organism: Sphingomonas sp. a1
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2017-10-04 Classification: STRUCTURAL PROTEIN Ligands: GOL, CU |
 |
Structure Of Alginate-Binding Protein Algq2 In Complex With An Alginate Pentasaccharide
Organism: Sphingomonas sp.
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2017-08-09 Classification: SUGAR BINDING PROTEIN Ligands: CA |
 |
Structure Of Alginate-Binding Protein Algq2 In Complex With An Alginate Trisaccharide
Organism: Sphingomonas sp.
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2017-08-09 Classification: SUGAR BINDING PROTEIN Ligands: CA, PEG, CL |
 |
Crystal Structure Of Solute-Binding Protein Complexed With Unsaturated Chondroitin Disaccharide With A Sulfate Group At C-4 Position Of Galnac
Organism: Streptobacillus moniliformis dsm 12112
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2017-07-26 Classification: SUGAR BINDING PROTEIN Ligands: CA, EDO |
 |
Crystal Structure Of Solute-Binding Protein Complexed With Sulfate Group-Free Unsaturated Chondroitin Disaccharide
Organism: Streptobacillus moniliformis strain dsm 12112
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2017-07-19 Classification: SUGAR BINDING PROTEIN Ligands: CA, MES, EDO |
 |
Crystal Structure Of Solute-Binding Protein Complexed With Unsaturated Chondroitin Disaccharide With A Sulfate Group At C-6 Position Of Galnac
Organism: Streptobacillus moniliformis dsm 12112
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2017-07-19 Classification: TRANSPORT PROTEIN Ligands: CA |
 |
Organism: Streptobacillus moniliformis dsm 12112
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2017-07-19 Classification: SUGAR BINDING PROTEIN Ligands: CA, EDO, SO4, TAR |