Search Count: 42
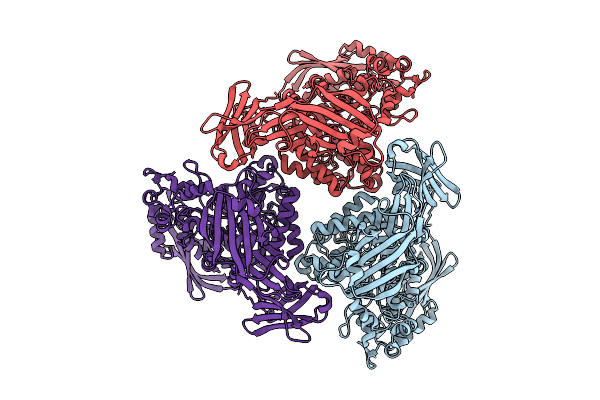 |
Active Conformation Of A Redox-Regulated Glycoside Hydrolase (Capgh2B) From The Gh2 Family
|
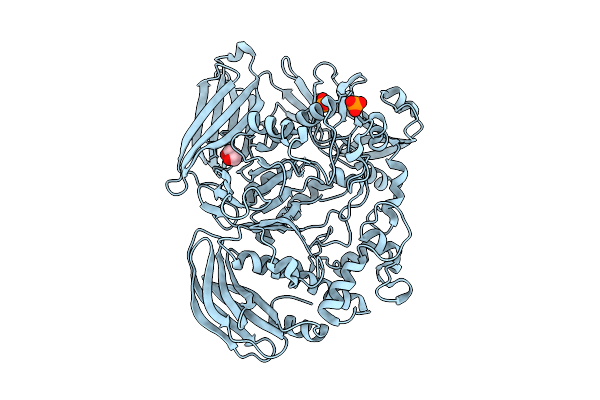 |
Crystal Structure Of The Inactive Conformation Of A Glycoside Hydrolase (Capgh2B) From The Gh2 Family In The Space Group I213 At 2.05 A
Organism: Metagenome
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: HYDROLASE Ligands: PO4, GOL |
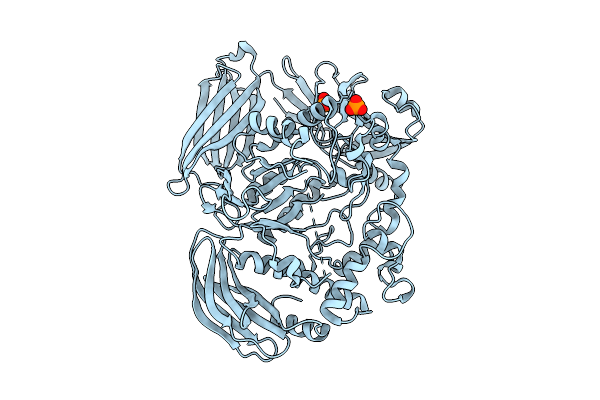 |
Crystal Structure Of The Inactive Conformation Of A Glycoside Hydrolase (Capgh2B - E553Q Mutant) From The Gh2 Family In The Space Group I213 At 2.6 A
Organism: Metagenome
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: HYDROLASE Ligands: PO4 |
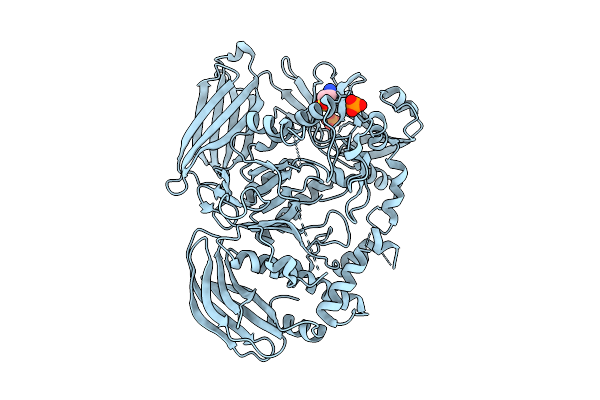 |
Crystal Structure Of The Inactive Conformation Of A Glycoside Hydrolase (Capgh2B) From The Gh2 Family In The Space Group I213 At 2.75 A
Organism: Metagenome
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: HYDROLASE Ligands: PO4, TAU |
 |
Crystal Structure Of The Inactive Conformation Of A Glycoside Hydrolase (Capgh2B) From The Gh2 Family In The Space Group R3 At 2.45 A
|
 |
Crystal Structure Of The Inactive Conformation Of A Glycoside Hydrolase (Capgh2B) From The Gh2 Family In The Space Group I212121 At 2.65 A
Organism: Metagenome
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: HYDROLASE Ligands: PO4, PEG, PG4 |
 |
Crystal Structure Of The Inactive Conformation Of A Glycoside Hydrolase (Capgh2B - E553Q Mutant) From The Gh2 Family In The Space Group P3121 At 3.05 A
Organism: Metagenome
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: HYDROLASE Ligands: PO4, ACT, MLI |
 |
Crystal Structure Of The Inactive Conformation Of A Glycoside Hydrolase (Capgh2B) From The Gh2 Family In The Space Group P1 At 2.40 A
Organism: Metagenome
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: HYDROLASE Ligands: PO4, GOL |
 |
Crystal Structure Of The Inactive Conformation Of A Glycoside Hydrolase (Capgh2B) From The Gh2 Family In The Space Group P1 At 2.15 A
Organism: Metagenome
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: HYDROLASE Ligands: PO4, GOL, ACT |
 |
Crystal Structure Of The Inactive Conformation Of A Glycoside Hydrolase (Capgh2B - E553Q Mutant) From The Gh2 Family In The Space Group P1 At 2.25 A
Organism: Metagenome
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: HYDROLASE Ligands: PO4, EDO |
 |
Crystal Structure Of The Inactive Conformation Of A Glycoside Hydrolase (Capgh2B - E465A Mutant) From The Gh2 Family In The Space Group P1 At 3.1 A
Organism: Metagenome
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: HYDROLASE Ligands: PO4, EDO |
 |
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-10-23 Classification: HYDROLASE Ligands: PEG, PGE, GOL, MG, NA, EPE, PO4 |
 |
Crystal Structure Of Capgh16_3 Enzyme Retrieved From Capybara Gut Metagenome
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2024-10-23 Classification: HYDROLASE Ligands: PO4, CA |
 |
Organism: Gut metagenome
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2024-10-23 Classification: HYDROLASE Ligands: GOL, PO4, PEG, PGE |
 |
Crystal Structure Of A Novel Gh5 Enzyme Retrieved From Capybara Gut Metagenome
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2022-11-02 Classification: HYDROLASE Ligands: 1PG, ACT, NA, CL |
 |
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:2.48 Å Release Date: 2021-09-08 Classification: HYDROLASE Ligands: GOL |
 |
Crystal Structure Of A Beta-Helix Domain Retrieved From Capybara Gut Metagenome
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2021-09-08 Classification: UNKNOWN FUNCTION Ligands: CA |
 |
Crystal Structure Of A Gh128 (Subgroup I) Endo-Beta-1,3-Glucanase From Amycolatopsis Mediterranei (Amgh128_I)
Organism: Amycolatopsis mediterranei
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2020-05-20 Classification: HYDROLASE Ligands: GOL, NA |
 |
Crystal Structure Of A Gh128 (Subgroup I) Endo-Beta-1,3-Glucanase From Amycolatopsis Mediterranei (Amgh128_I) In Complex With Laminaritriose
Organism: Amycolatopsis mediterranei
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2020-05-20 Classification: HYDROLASE |
 |
Crystal Structure Of A Gh128 (Subgroup I) Endo-Beta-1,3-Glucanase (E199A Mutant) From Amycolatopsis Mediterranei (Amgh128_I) In Complex With Laminaripentaose
Organism: Amycolatopsis mediterranei
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2020-05-20 Classification: HYDROLASE Ligands: BGC, ZN, PEG |

