Search Count: 23
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2025-02-26 Classification: METAL BINDING PROTEIN Ligands: PGD, 6MO, PEG, PGE, MLT, 1PE |
 |
Discovery Of The Lanthipeptide Curvocidin And Structural Insights Into Its Trifunctional Synthetase Cuvl
Organism: Thermomonospora curvata
Method: X-RAY DIFFRACTION Resolution:2.68 Å Release Date: 2023-06-14 Classification: BIOSYNTHETIC PROTEIN Ligands: PO4, NO3 |
 |
Discovery Of The Lanthipeptide Curvocidin And Structural Insights Into Its Trifunctional Synthetase Cuvl
Organism: Thermomonospora curvata
Method: X-RAY DIFFRACTION Resolution:2.87 Å Release Date: 2023-06-14 Classification: BIOSYNTHETIC PROTEIN Ligands: MPD, ANP, MG |
 |
Organism: Bacillus licheniformis
Method: X-RAY DIFFRACTION Resolution:2.82 Å Release Date: 2022-08-10 Classification: ANTIMICROBIAL PROTEIN Ligands: ZN, ANP, GOL, MG |
 |
Organism: Methanocaldococcus jannaschii (strain atcc 43067 / dsm 2661 / jal-1 / jcm 10045 / nbrc 100440)
Method: X-RAY DIFFRACTION Resolution:2.43 Å Release Date: 2019-01-16 Classification: LIGASE Ligands: 96Z, GOL, CA, PG4 |
 |
In Silico-Powered Specific Incorporation Of Photocaged Dopa At Multiple Protein Sites
Organism: Methanocaldococcus jannaschii (strain atcc 43067 / dsm 2661 / jal-1 / jcm 10045 / nbrc 100440)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2017-09-13 Classification: LIGASE Ligands: 1PE, CA, CL, BUU |
 |
Substrate Bound Sn-Glycerol-3-Phosphate Binding Periplasmic Protein Ugpb From Escherichia Coli
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2012-10-17 Classification: DIESTER-BINDING PROTEIN Ligands: G3P, GOL, CD, CO, MG |
 |
Organism: Clostridium scatologenes
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2011-09-21 Classification: LYASE Ligands: SF4 |
 |
Organism: Clostridium scatologenes
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2011-09-21 Classification: LYASE Ligands: 4HP, SF4 |
 |
Crystal Structure Of The Human Transcription Elongation Factor Dsif, Hspt4/Hspt5 (176-273)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2009-12-08 Classification: TRANSCRIPTION Ligands: BME, ZN |
 |
Organism: Silicibacter pomeroyi
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2007-04-10 Classification: UNKNOWN FUNCTION Ligands: IOD, HEM |
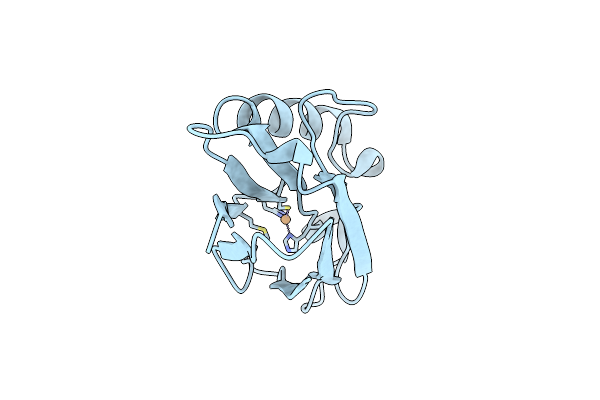 |
Structure Of Cu(Ii)Azurin With The Metal-Binding Loop Sequence "Ctfpghsalm" Replaced With "Ctphpm"
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2006-04-11 Classification: ELECTRON TRANSPORT Ligands: CU |
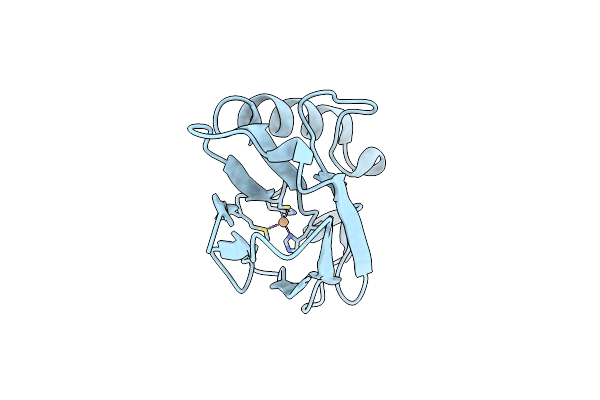 |
Structure Of Cu(I)Azurin At Ph 6, With The Metal-Binding Loop Sequence "Ctfpghsalm" Replaced With "Ctphpm"
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2006-04-11 Classification: ELECTRON TRANSPORT Ligands: CU1 |
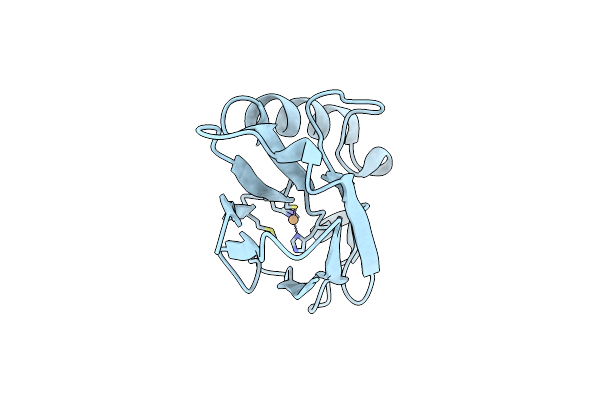 |
Structure Of Cu(I)Azurin, Ph8, With The Metal-Binding Loop Sequence "Ctfpghsalm" Replaced With "Ctphpm"
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2006-04-11 Classification: ELECTRON TRANSPORT Ligands: CU1 |
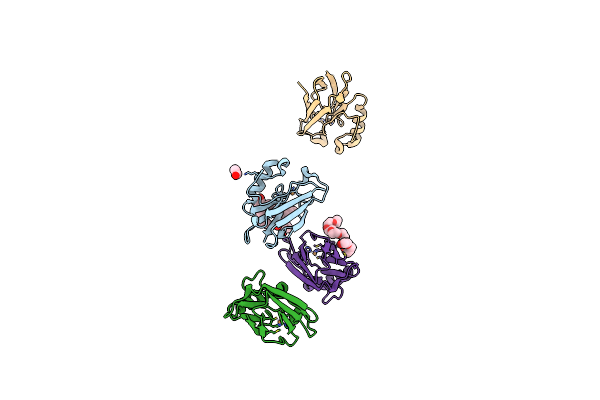 |
Structure Of Cu(Ii)Azurin With The Metal-Binding Loop Sequence "Ctfpghsalm" Replaced With "Ctphpfm"
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.61 Å Release Date: 2006-04-11 Classification: ELECTRON TRANSPORT Ligands: CU, 2PE, EOH, PEG |
 |
2-Oxoquinoline 8-Monooxygenase Component: Active Site Modulation By Rieske-[2Fe-2S] Center Oxidation/Reduction
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2005-05-24 Classification: OXIDOREDUCTASE Ligands: FE, FES |
 |
2-Oxoquinoline 8-Monooxygenase Component: Active Site Modulation By Rieske-[2Fe-2S] Center Oxidation/Reduction
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2005-05-24 Classification: OXIDOREDUCTASE Ligands: FE, FES |
 |
2-Oxoquinoline 8-Monooxygenase Component: Active Site Modulation By Rieske-[2Fe-2S] Center Oxidation/Reduction
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2005-05-24 Classification: OXIDOREDUCTASE Ligands: FE, FES, OCH |
 |
Nad+-Dependent (R)-2-Hydroxyglutarate Dehydrogenase From Acidaminococcus Fermentans
Organism: Acidaminococcus fermentans
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2005-03-08 Classification: OXIDOREDUCTASE |
 |
Crystal Structure Of 4-Hydroxybutyryl-Coa Dehydratase From Clostridium Aminobutyricum: Radical Catalysis Involving A [4Fe-4S] Cluster And Flavin
Organism: Clostridium aminobutyricum
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2004-12-21 Classification: LYASE, ISOMERASE Ligands: SF4, FAD |

