Search Count: 18
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: MOTOR PROTEIN Ligands: ADP, VO4, MG, DMS, EDO, A1IGE, CL |
 |
Structure Of A Pseudomurein Peptide Ligase Type E From Methanothermobacter Thermautotrophicus
Organism: Methanothermobacter thermautotrophicus
Method: X-RAY DIFFRACTION Resolution:2.91 Å Release Date: 2022-10-12 Classification: LIGASE |
 |
Structure Of A Pseudomurein Peptide Ligase Type E From Methanothermus Fervidus
Organism: Methanothermus fervidus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-10-12 Classification: LIGASE Ligands: UDP, SO4 |
 |
Apo Structure Of A Pseudomurein Peptide Ligase Type E From Methanothermus Fervidus
Organism: Methanothermus fervidus (strain atcc 43054 / dsm 2088 / jcm 10308 / v24 s)
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2021-09-01 Classification: LIGASE Ligands: MG, SO4 |
 |
Structure Of A Pseudomurein Peptide Ligase Type E From Methanothermus Fervidus
Organism: Methanothermus fervidus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2021-08-11 Classification: LIGASE Ligands: PO4, MG, EDO, SO4, UDP |
 |
Crystal Structure Of A Truncated Lsd1:Corest In The Presence Of An Lsd1-Nt Peptide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2021-06-30 Classification: OXIDOREDUCTASE Ligands: FAD |
 |
Structure Of A Stable Interstrand Dna Crosslink Involving An Da Amino Group And An Abasic Site
|
 |
Structure Of A Pseudomurein Peptide Ligase Type C From Methanothermus Fervidus
Organism: Methanothermus fervidus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2021-02-10 Classification: LIGASE Ligands: SO4, ZN, GOL, AAE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2016-04-27 Classification: Oxidoreductase/Transcription/RNA Ligands: FAD, SO4, GOL |
 |
Organism: Methanocaldococcus jannaschii dsm 2661
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2015-07-22 Classification: OXIDOREDUCTASE Ligands: NDP, K, ZN, EDO, NA |
 |
Crystal Structure Of The Methanocaldococcus Jannaschii G1Pdh With Nadph And Dhap
Organism: Methanocaldococcus jannaschii
Method: X-RAY DIFFRACTION Resolution:2.23 Å Release Date: 2015-07-22 Classification: OXIDOREDUCTASE Ligands: NDP, K, ZN, 13P, EDO, 1GP |
 |
Organism: Methanocaldococcus jannaschii
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2015-07-22 Classification: OXIDOREDUCTASE Ligands: MG, ZN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2003-05-13 Classification: IMMUNE RESPONSE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2003-05-13 Classification: IMMUNE SYSTEM |
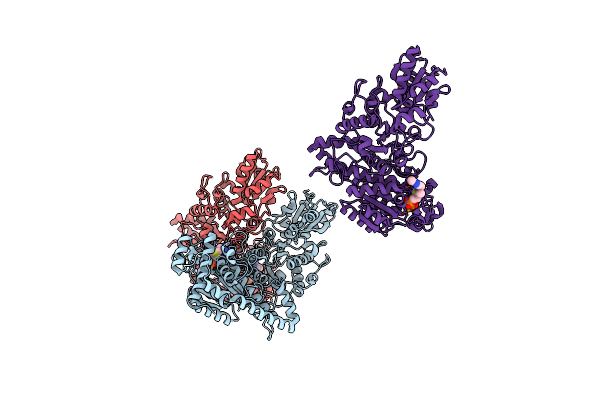 |
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2003-02-15 Classification: TRANSFERASE Ligands: MG, TPP |
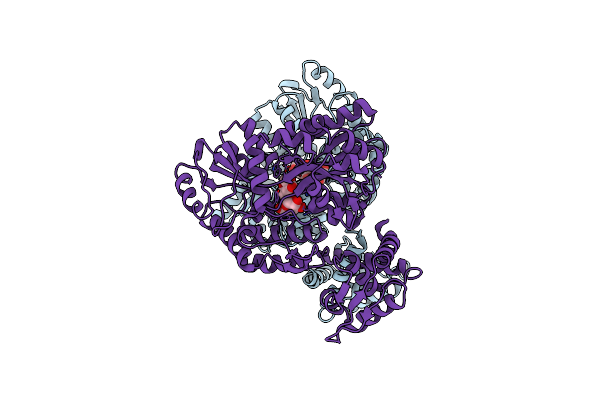 |
Organism: Escherichia coli, Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2001-06-06 Classification: Transcription/Sugar Binding Protein Ligands: GLC |
 |
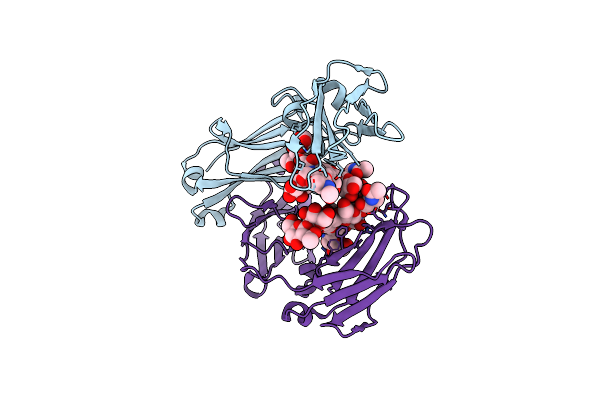
Crystal Structure Of The Neonatal Fc Receptor Complexed With A Heterodimeric Fc
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2001-02-14 Classification: IMMUNE SYSTEM Ligands: NAG, CYS |
 |
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2001-02-14 Classification: IMMUNE SYSTEM |

