Search Count: 15
 |
Crystal Structure Of A Far-Red Cyanobacteriochrome Photoreceptor At Room Temperature
Organism: Anabaena cylindrica (strain atcc 27899 / pcc 7122)
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2020-11-04 Classification: SIGNALING PROTEIN Ligands: CYC |
 |
Organism: Anabaena cylindrica (strain atcc 27899 / pcc 7122)
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2020-11-04 Classification: SIGNALING PROTEIN Ligands: CYC |
 |
Organism: Nostoc punctiforme (strain atcc 29133 / pcc 73102)
Method: SOLUTION NMR Release Date: 2018-04-18 Classification: SIGNALING PROTEIN Ligands: CYC |
 |
Organism: Nostoc punctiforme pcc 73102
Method: SOLUTION NMR Release Date: 2018-04-18 Classification: SIGNALING PROTEIN Ligands: CYC |
 |
Organism: Synechocystis sp.
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2010-10-06 Classification: OXIDOREDUCTASE Ligands: EDO, BLA |
 |
Organism: Synechocystis sp.
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2010-10-06 Classification: OXIDOREDUCTASE Ligands: EDO, BLA |
 |
Organism: Synechocystis sp.
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2009-10-13 Classification: OXIDOREDUCTASE Ligands: EDO, BLA |
 |
Organism: Synechocystis sp.
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2009-10-13 Classification: OXIDOREDUCTASE Ligands: EDO, BLA |
 |
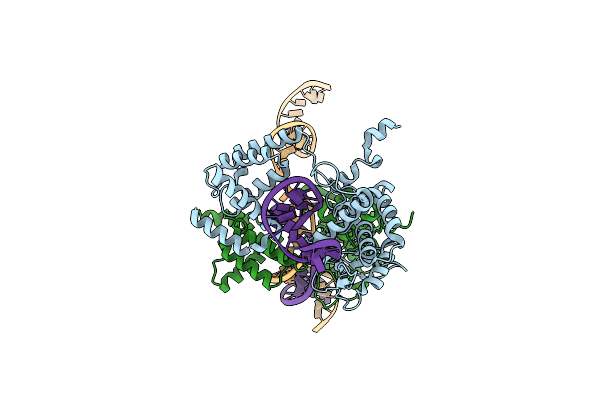
Basis For A Switch In Substrate Specificity: Crystal Structure Of Selected Variant Of Cre Site-Specific Recombinase, Alshg Bound To The Engineered Recognition Site Loxm7
Organism: Escherichia phage p1, Escherichia virus p1
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2004-02-17 Classification: RECOMBINATION/DNA |
 |
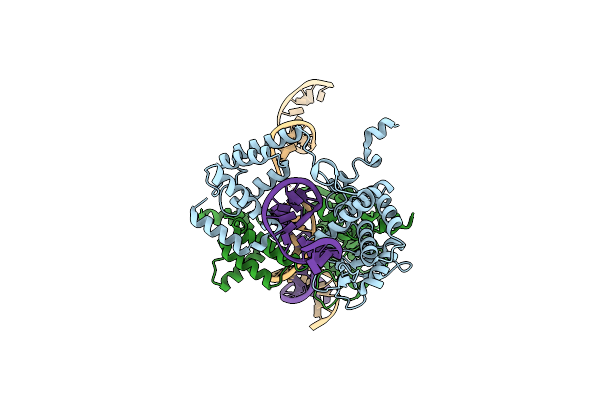
Basis For A Switch In Substrate Specificity: Crystal Structure Of Selected Variant Of Cre Site-Specific Recombinase, Lnsgg Bound To The Engineered Recognition Site Loxm7
Organism: Escherichia phage p1, Escherichia virus p1
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2004-02-17 Classification: RECOMBINATION/DNA |
 |
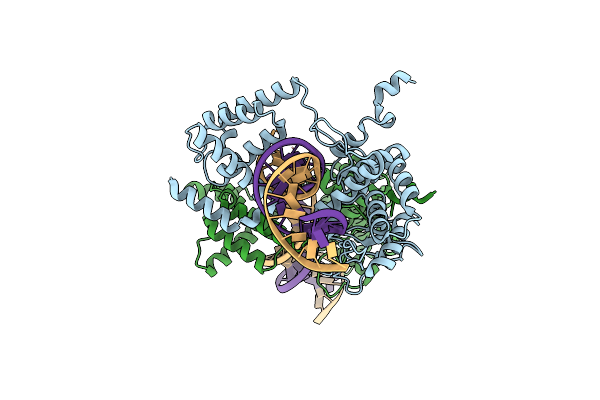
Basis For A Switch In Substrate Specificity: Crystal Structure Of Selected Variant Of Cre Site-Specific Recombinase, Lnsgg Bound To The Loxp (Wildtype) Recognition Site
Organism: Enterobacteria phage p1
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2004-02-17 Classification: RECOMBINATION/DNA |
 |
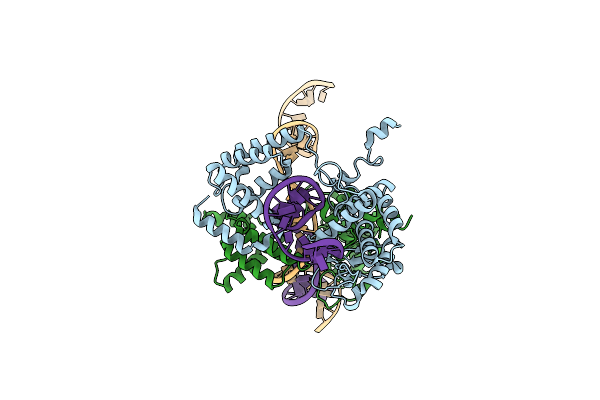
Crystal Structure Of Cre Site-Specific Recombinase Complexed With A Mutant Dna Substrate, Loxp-A8/T27
Organism: Enterobacteria phage p1
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2003-07-08 Classification: HYDROLASE, LIGASE/DNA |
 |
Organism: Enterobacteria phage p1
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2002-06-07 Classification: HYDROLASE, LIGASE/DNA |
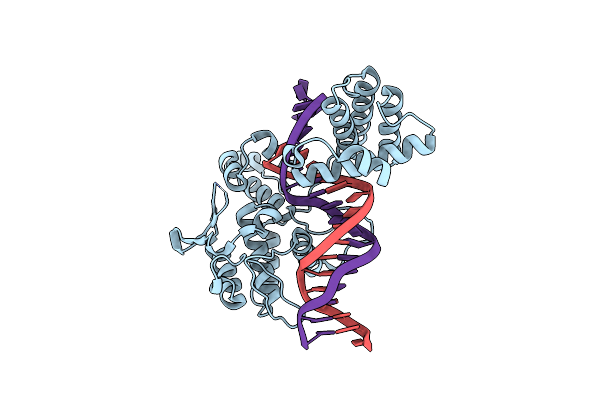 |
Organism: Enterobacteria phage p1
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2001-10-19 Classification: HYDROLASE, LIGASE/DNA |
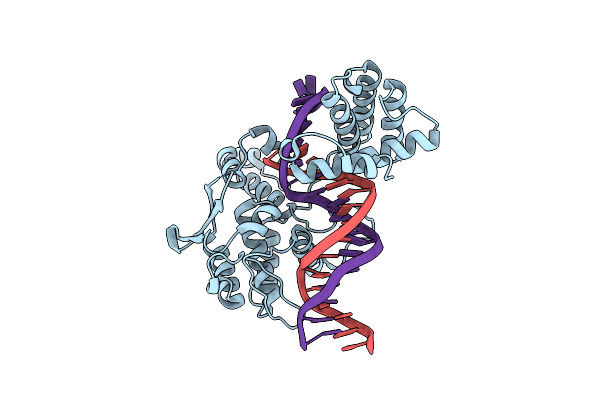 |
Organism: Enterobacteria phage p1
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2001-10-12 Classification: HYDROLASE, LIGASE/DNA |

