Search Count: 175
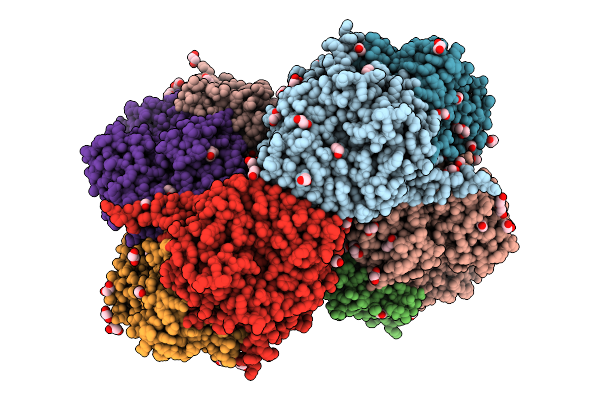 |
Organism: Azotobacter vinelandii dj
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: METAL BINDING PROTEIN Ligands: SF4, S5Q, PEG, EDO, 1PE, PGE, MG, PG4 |
 |
Solution Nmr Structure Of A Peptide Encompassing Residues 967-991 Of The Human Formin Inf2
|
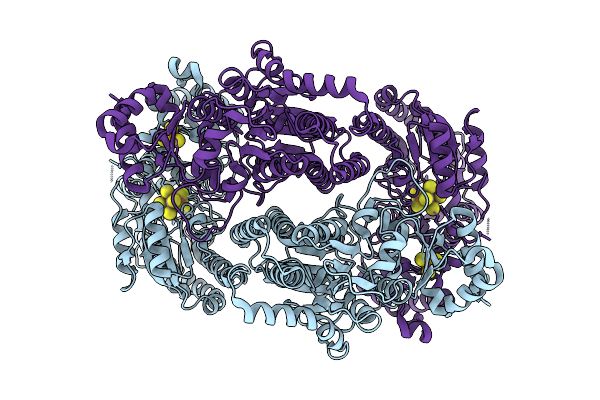 |
Organism: Geobacter metallireducens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: METAL BINDING PROTEIN Ligands: SF4, S5Q |
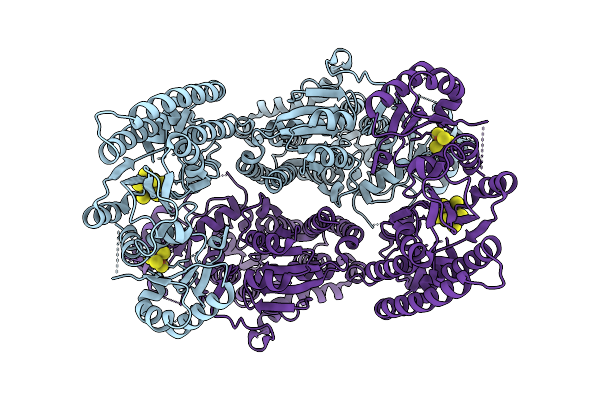 |
Organism: Geobacter metallireducens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: METAL BINDING PROTEIN Ligands: SF4, S5Q |
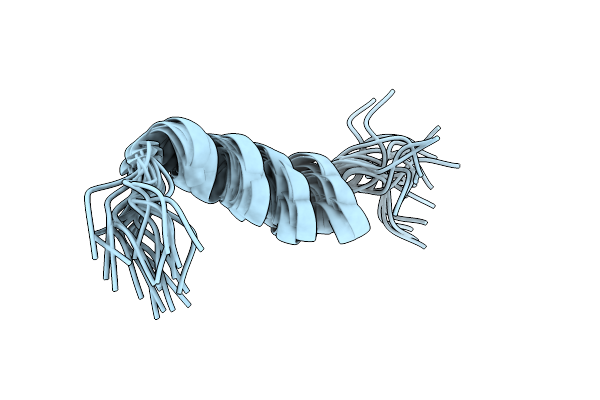 |
Solution Nmr Structure Of A Peptide Encompassing Residues 967-991 Of The Human Formin Inf2
|
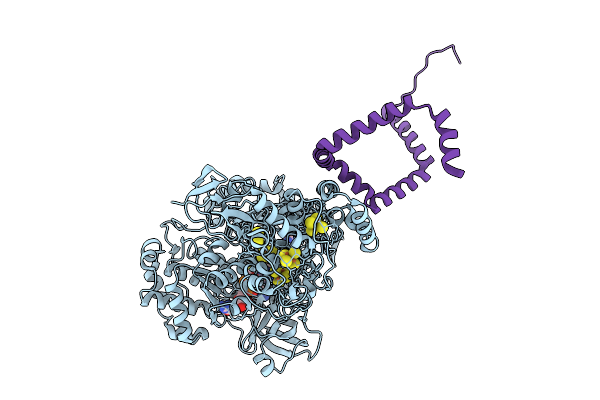 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Release Date: 2025-09-10 Classification: ELECTRON TRANSPORT Ligands: FES, SF4, MGD, 4MO |
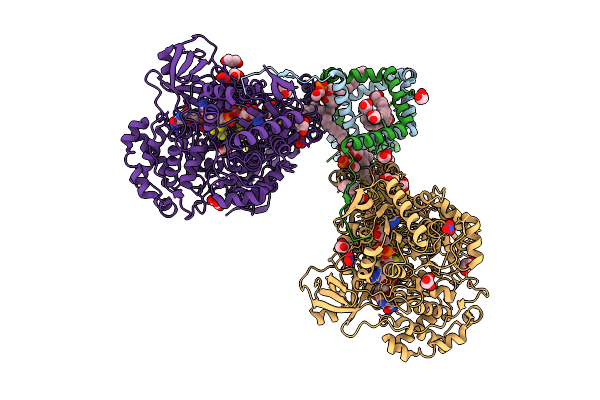 |
Crystal Structure Of Molybdenum Bispyranopterin Guanine Dinucleotide Formate Dehydrogenases Force1 From Bacillus Subtilis
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: OXIDOREDUCTASE Ligands: MYR, GOL, LHG, FES, SF4, MGD, 4MO, MQ7, H2S, MLI, NO3, NA |
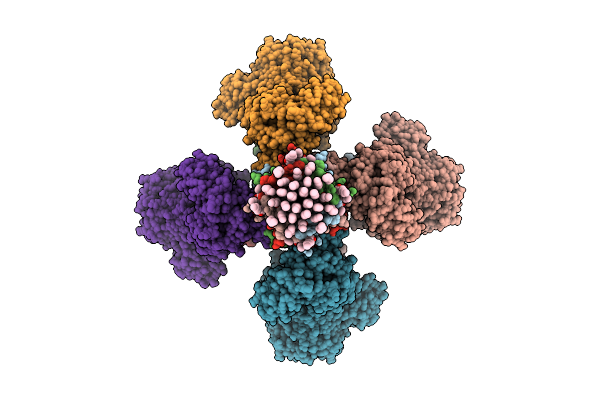 |
Cryoem Structure Of Molybdenum Bispyranopterin Guanine Dinucleotide Formate Dehydrogenases Force1 From Bacillus Subtilis
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: ELECTRON MICROSCOPY Release Date: 2025-08-06 Classification: OXIDOREDUCTASE Ligands: 11A, A1H2V, SHV, KNA, MYR, FES, SF4, MGD, 4MO, H2S, MQ7 |
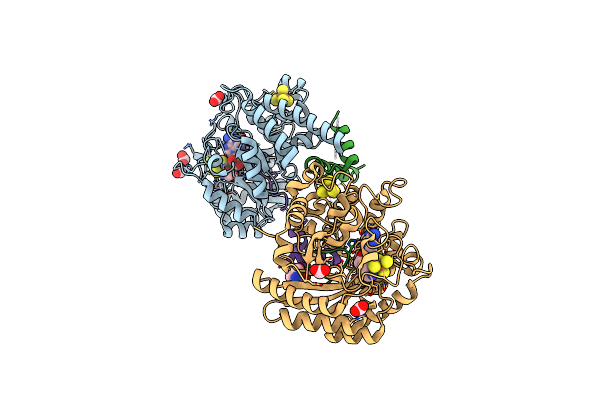 |
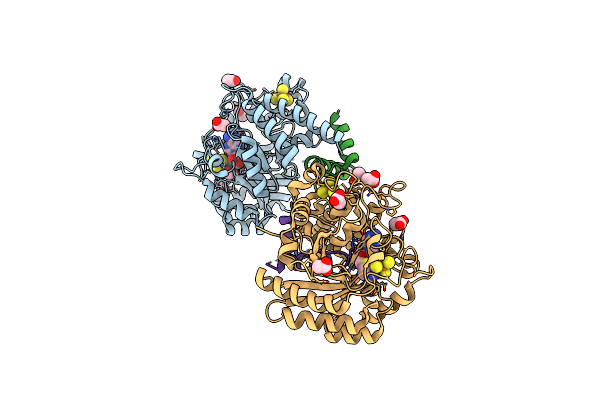
Crystal Structure Of The Complex Formed Between The Radical Sam Protein Chlb And The Leader Region Of Its Precursor Substrate Chla
Organism: Fischerella
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: OXIDOREDUCTASE Ligands: SF4, FMT, SAH, TRS, ZN |
 |
Crystal Structure Of The Complex Formed Between The Radical Sam Protein Chlb And The R3A Mutant Of Chla
Organism: Fischerella
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: OXIDOREDUCTASE Ligands: SF4, SAH, ACT |
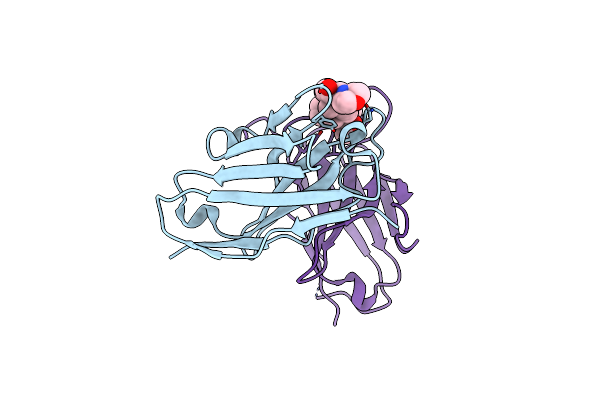 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2025-02-12 Classification: IMMUNE SYSTEM Ligands: RBR |
 |
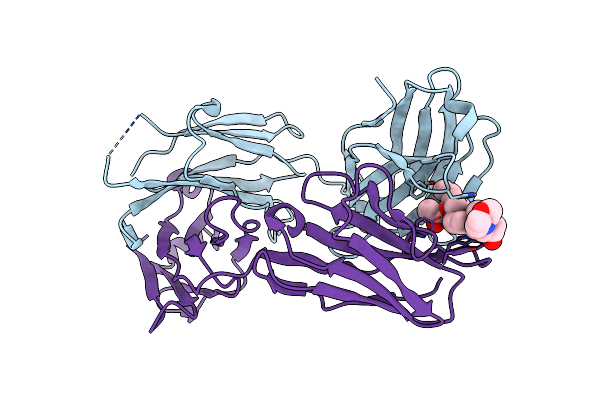
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2025-02-12 Classification: IMMUNE SYSTEM Ligands: RBR |
 |
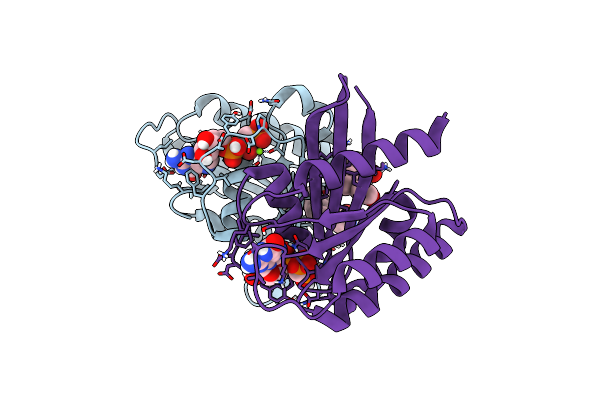
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.37 Å Release Date: 2025-02-12 Classification: IMMUNE SYSTEM Ligands: RBR |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.16 Å Release Date: 2024-10-02 Classification: ONCOPROTEIN Ligands: GCP, YLE, MG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.31 Å Release Date: 2024-10-02 Classification: ONCOPROTEIN Ligands: GCP, MG, YFJ |
 |
Organism: Christiangramia forsetii kt0803
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-09-18 Classification: SUGAR BINDING PROTEIN |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2024-09-18 Classification: IMMUNE SYSTEM Ligands: RBR |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2024-09-18 Classification: IMMUNE SYSTEM Ligands: RBR, EPE |
 |
Solution Nmr Structure Of A Peptide Encompassing Residues 2-19 Of The Human Formin Inf2
|
 |
Solution Nmr Structure Of A Peptide Encompassing Residues 2-36 Of The Human Formin Inf2
|

