Search Count: 450
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: A1IAT |
 |
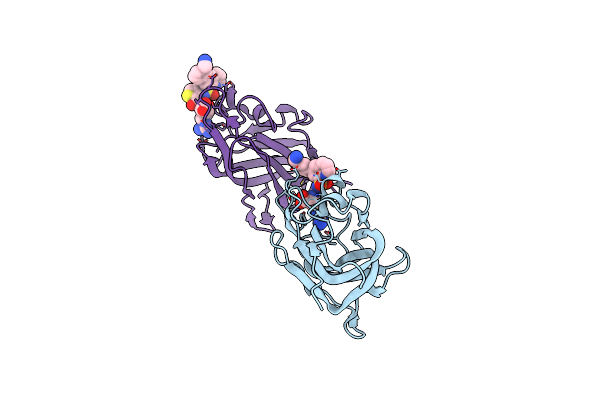
Single-Particle Cryo-Em Of Mycoplasma Pneumoniae Adhesin P1 Complexed With The Anti-Adhesive Fab Fragment.
Organism: Mycoplasmoides pneumoniae m129, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: CELL ADHESION |
 |
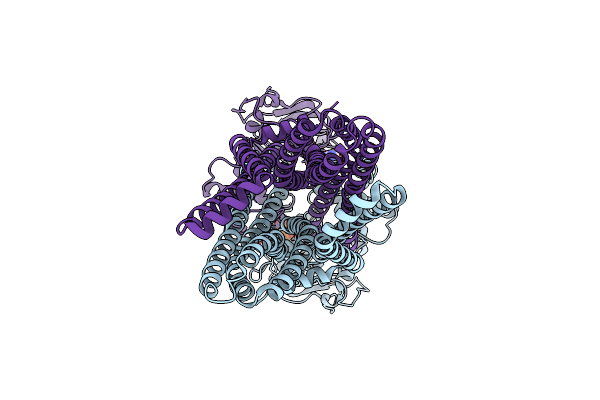
Organism: Bacillus subtilis
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: MEMBRANE PROTEIN Ligands: HT1 |
 |
Organism: Bacillus subtilis
Method: ELECTRON MICROSCOPY Release Date: 2025-01-29 Classification: TRANSPORT PROTEIN Ligands: ATP, MG |
 |
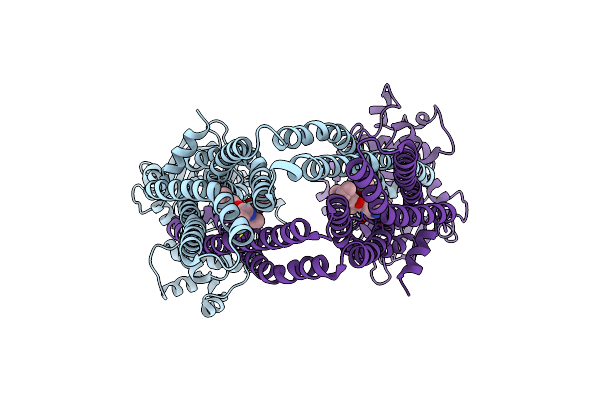
Organism: Bacillus subtilis
Method: ELECTRON MICROSCOPY Release Date: 2025-01-01 Classification: TRANSPORT PROTEIN |
 |
Organism: Bacillus subtilis
Method: ELECTRON MICROSCOPY Release Date: 2025-01-01 Classification: TRANSPORT PROTEIN Ligands: RHQ |
 |
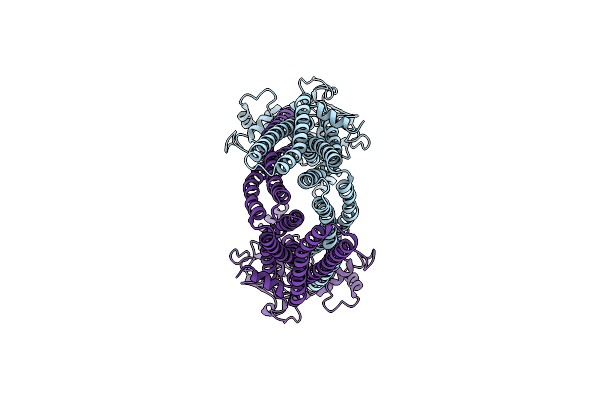
Organism: Bacillus subtilis
Method: ELECTRON MICROSCOPY Release Date: 2025-01-01 Classification: TRANSPORT PROTEIN Ligands: RHQ |
 |
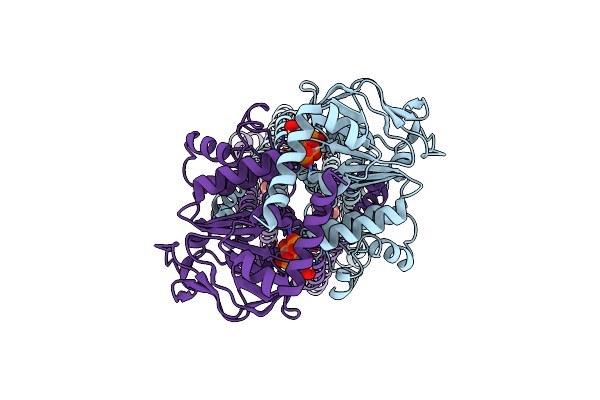
Organism: Bacillus subtilis
Method: ELECTRON MICROSCOPY Release Date: 2025-01-01 Classification: TRANSPORT PROTEIN |
 |
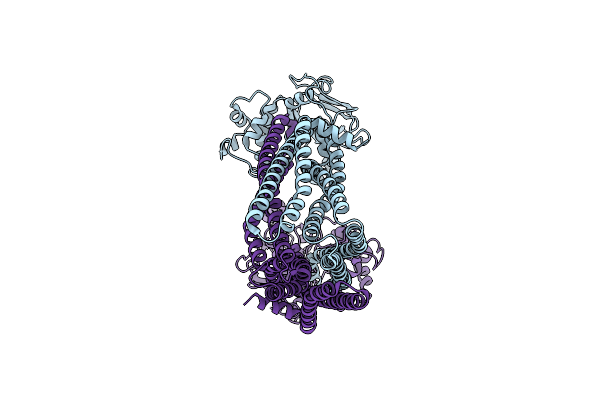
Organism: Bacillus subtilis
Method: ELECTRON MICROSCOPY Release Date: 2025-01-01 Classification: TRANSPORT PROTEIN Ligands: RHQ, ATP, MG |
 |
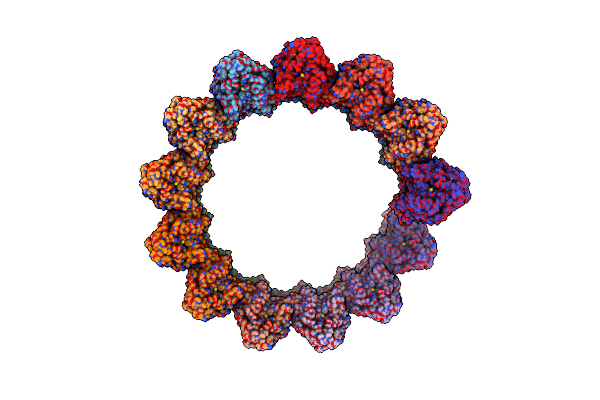
Organism: Bacillus subtilis
Method: ELECTRON MICROSCOPY Release Date: 2025-01-01 Classification: TRANSPORT PROTEIN |
 |
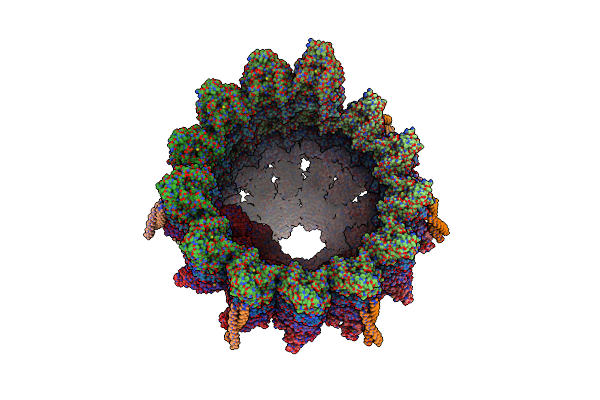
Structure Of The Native Microtubule Lattice Nucleated From The Yeast Spindle Pole Body
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2024-04-24 Classification: CELL CYCLE |
 |
Structure Of The Native Y-Tubulin Ring Complex (Yturc) Capping Microtubule Minus Ends At The Spindle Pole Body
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2024-04-24 Classification: CELL CYCLE Ligands: GTP, GDP |
 |
Structure Of The Y-Tubulin Small Complex (Ytusc) As Part Of The Native Y-Tubulin Ring Complex (Yturc) Capping Microtubule Minus Ends At The Spindle Pole Body
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2024-04-24 Classification: CELL CYCLE Ligands: GTP |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2024-04-03 Classification: CELL CYCLE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2024-02-07 Classification: HYDROLASE Ligands: MG, X5W |
 |
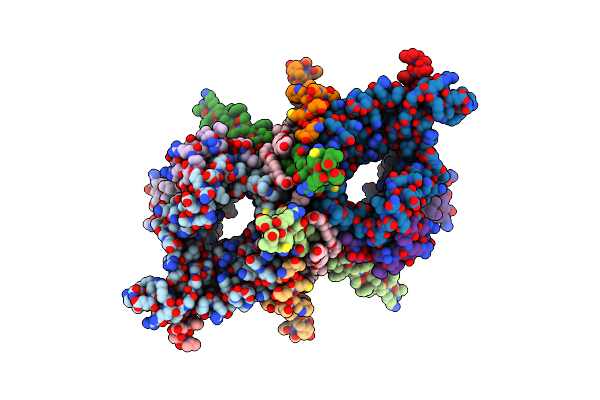
Crystal Structure Of A Muc1-Like Glycopeptide Containing The Unnatural L-4-Hydroxynorvaline In Complex With Scfv-Sm3
Organism: Mus musculus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2023-08-09 Classification: IMMUNE SYSTEM Ligands: EDO, A2G |
 |
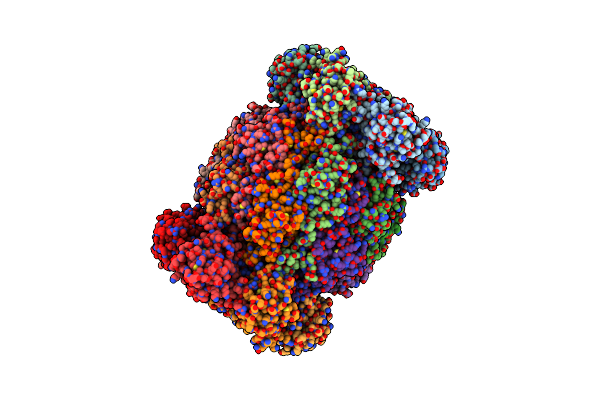
Cryo-Em Structure Of The Neurospora Crassa Tom Core Complex At 3.3 Angstrom
Organism: Neurospora crassa
Method: ELECTRON MICROSCOPY Release Date: 2023-08-09 Classification: MEMBRANE PROTEIN Ligands: DU0, PLC |
 |
Leishmania Tarentolae Proteasome 20S Subunit In Complex With 1-Benzyl-N-(3-(Cyclopropylcarbamoyl)Phenyl)-6-Oxo-1,6-Dihydropyridazine-3-Carboxamide
Organism: Leishmania tarentolae
Method: ELECTRON MICROSCOPY Resolution:2.59 Å Release Date: 2023-08-09 Classification: UNKNOWN FUNCTION Ligands: VYW |
 |
Crystal Structure Of Hla-B*15:01 In Complex With Spike Derived Peptide Nqklianaf From Oc43 Virus
Organism: Homo sapiens, Human coronavirus 229e
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2023-06-14 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: ACT |
 |
Crystal Structure Of Hla-B*15:01 In Complex With Spike Derived Peptide Nqklianqf From Sars-Cov-2 Virus
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2023-06-14 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: ACT, PEG |

