Search Count: 50
 |
X-Ray Crystal Structure Of Wild-Type Thermothelomyces Thermophilus Polysaccharide Monooxygenase 9E
Organism: Thermothelomyces thermophilus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-12-18 Classification: OXIDOREDUCTASE Ligands: CU, ACT, OXY, EDO |
 |
X-Ray Crystal Structure Of Y168F Variant Thermothelomyces Thermophilus Polysaccharide Monooxygenase 9E
Organism: Thermothelomyces thermophilus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-12-18 Classification: OXIDOREDUCTASE Ligands: CU, OXY, ACT, EDO |
 |
X-Ray Crystal Structure Of Y62W Thermothelomyces Thermophilus Polysaccharide Monooxygenase 9E
Organism: Thermothelomyces thermophilus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-12-18 Classification: OXIDOREDUCTASE Ligands: CU, EDO, ACY, OXY |
 |
X-Ray Crystal Structure Of Y62F Thermothelomyces Thermophilus Polysaccharide Monooxygenase 9E
Organism: Thermothelomyces thermophilus
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2024-12-18 Classification: OXIDOREDUCTASE Ligands: CU, EDO, OXY |
 |
X-Ray Crystal Structure Of H157Q Variant Thermothelomyces Thermophilus Polysaccharide Monooxygenase 9E
Organism: Thermothelomyces thermophilus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-12-04 Classification: OXIDOREDUCTASE Ligands: CU, OXY, EDO |
 |
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:1.14 Å Release Date: 2020-06-24 Classification: LYASE Ligands: NI |
 |
Crystal Structure Of The Hydroxyglutarate Synthase In Complex With 2-Oxoadipate From Pseudomonas Putida
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:1.42 Å Release Date: 2020-06-24 Classification: LYASE Ligands: NI, OOG |
 |
Crystal Structure Of The Hydroxyglutarate Synthase In Complex With 2-Oxoadipate From Oryza Sativa
Organism: Oryza sativa
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2020-06-24 Classification: LYASE Ligands: NI, OOG, SO4 |
 |
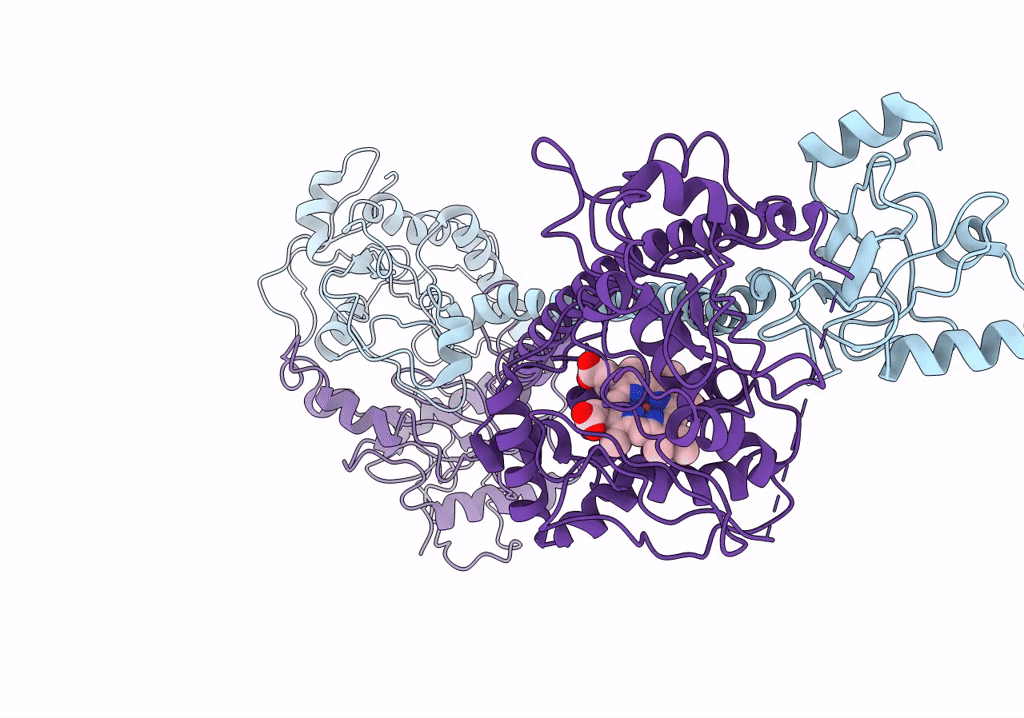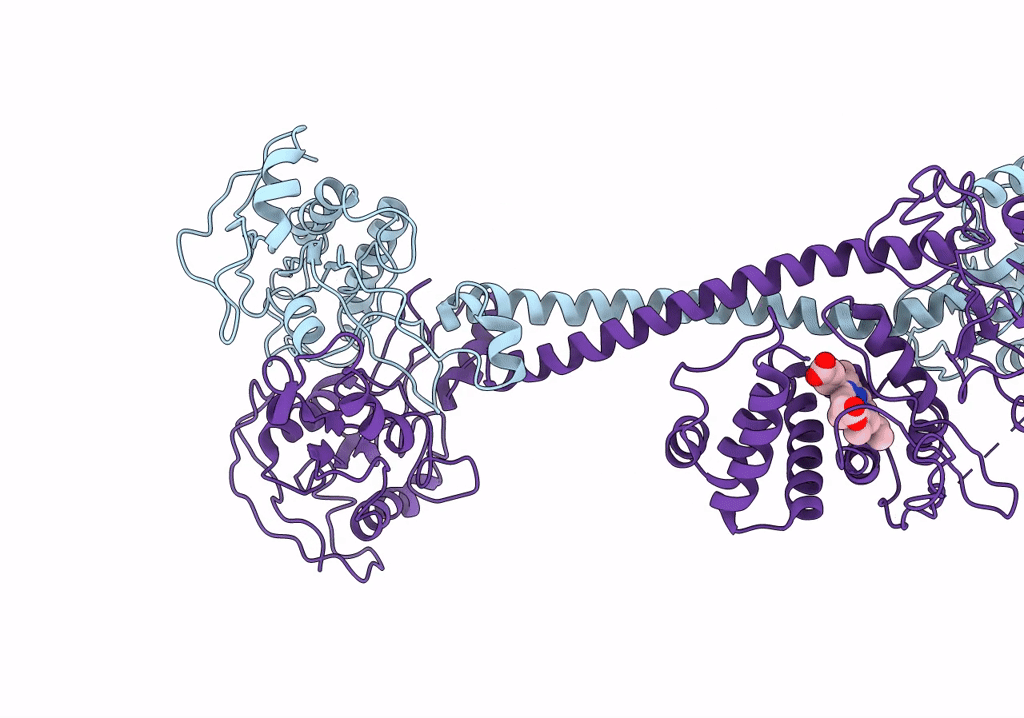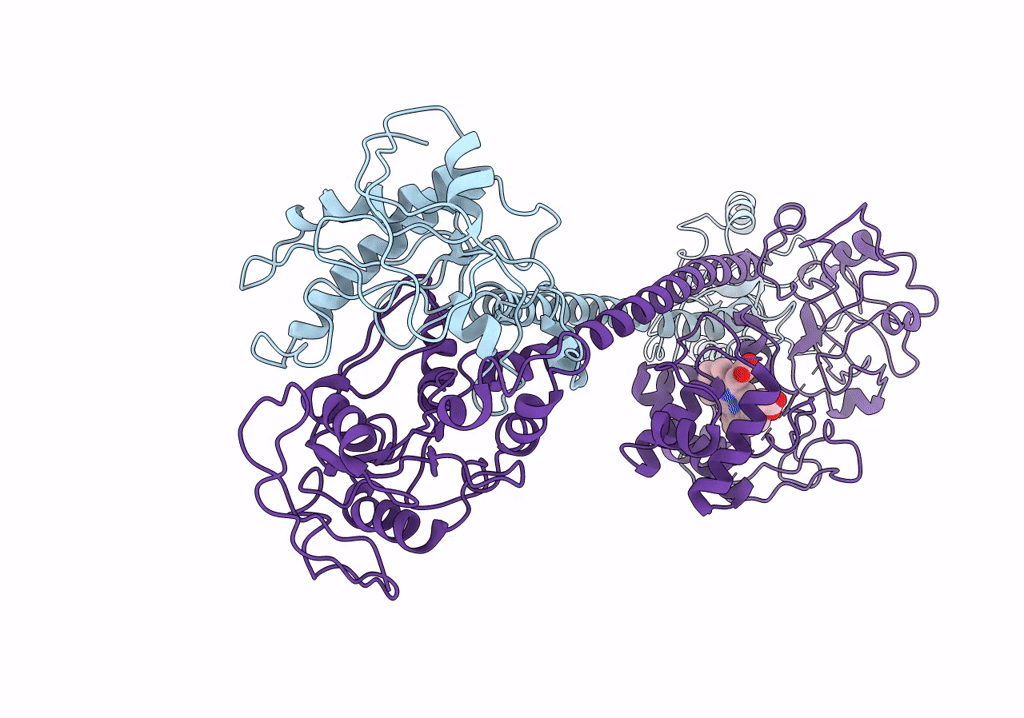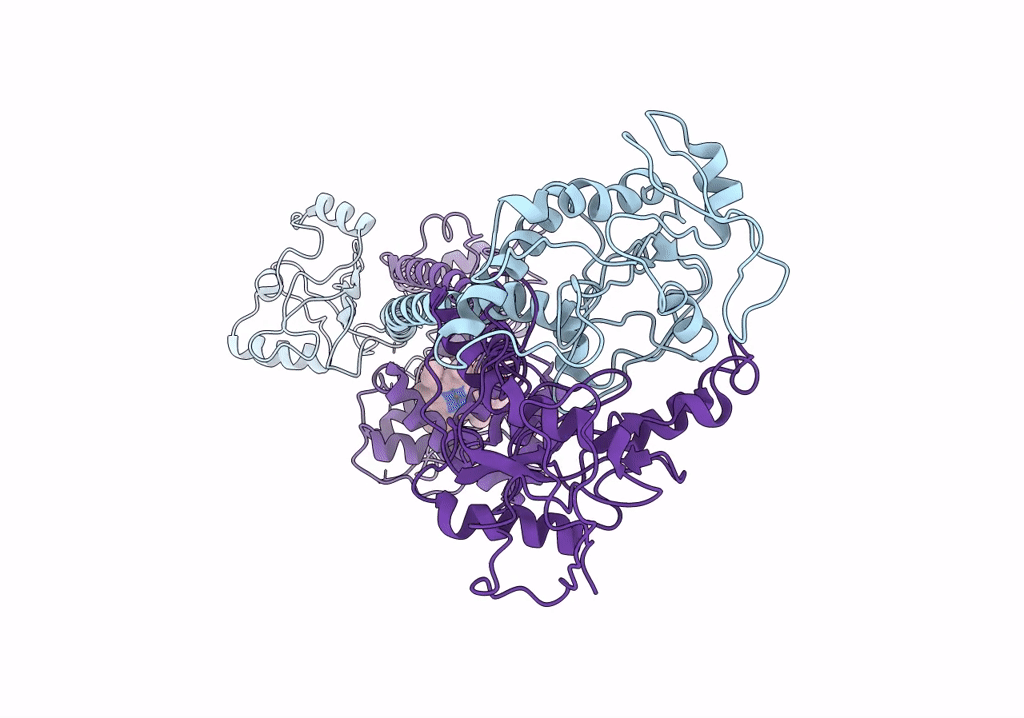
Organism: Manduca sexta
Method: ELECTRON MICROSCOPY Release Date: 2019-10-23 Classification: SIGNALING PROTEIN Ligands: HEM |
 |
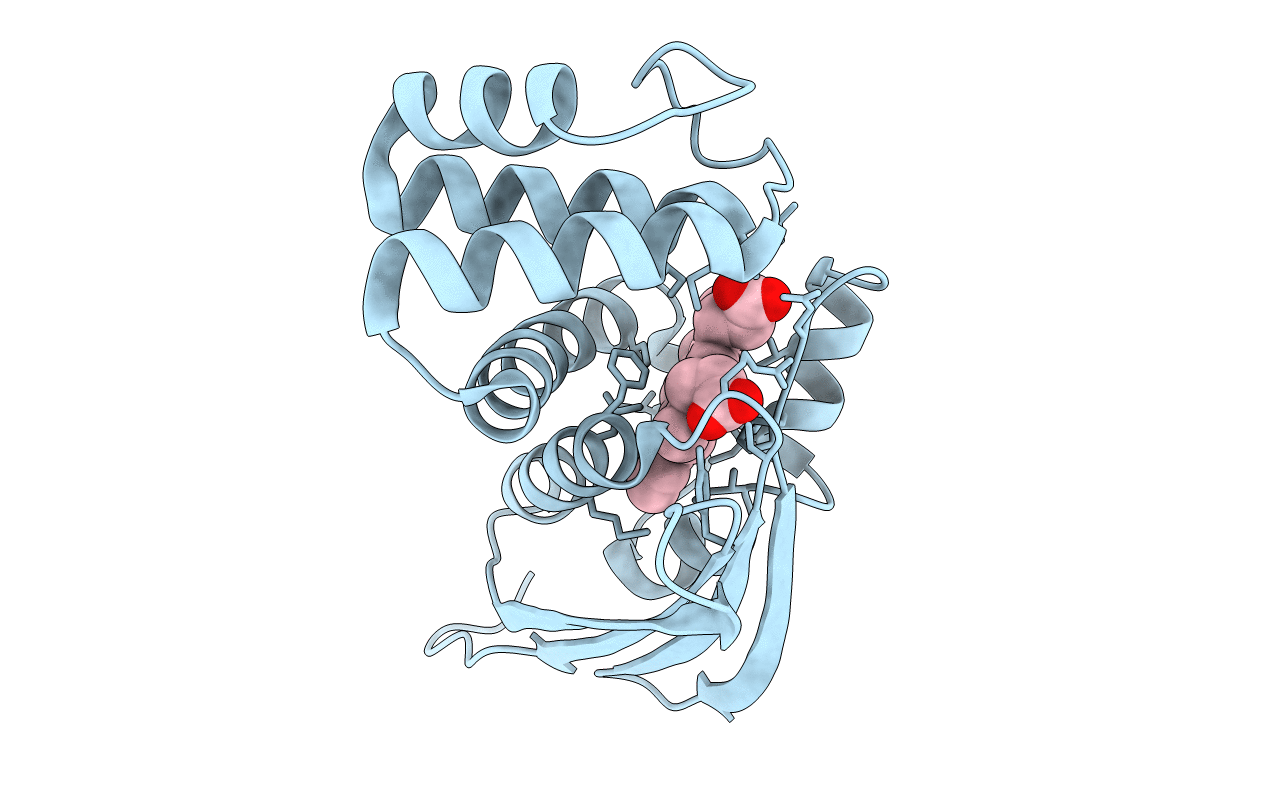
Organism: Manduca sexta
Method: ELECTRON MICROSCOPY Release Date: 2019-10-23 Classification: SIGNALING PROTEIN Ligands: HEM |
 |
Organism: Kordia algicida ot-1
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2018-05-30 Classification: SIGNALING PROTEIN Ligands: HEM |
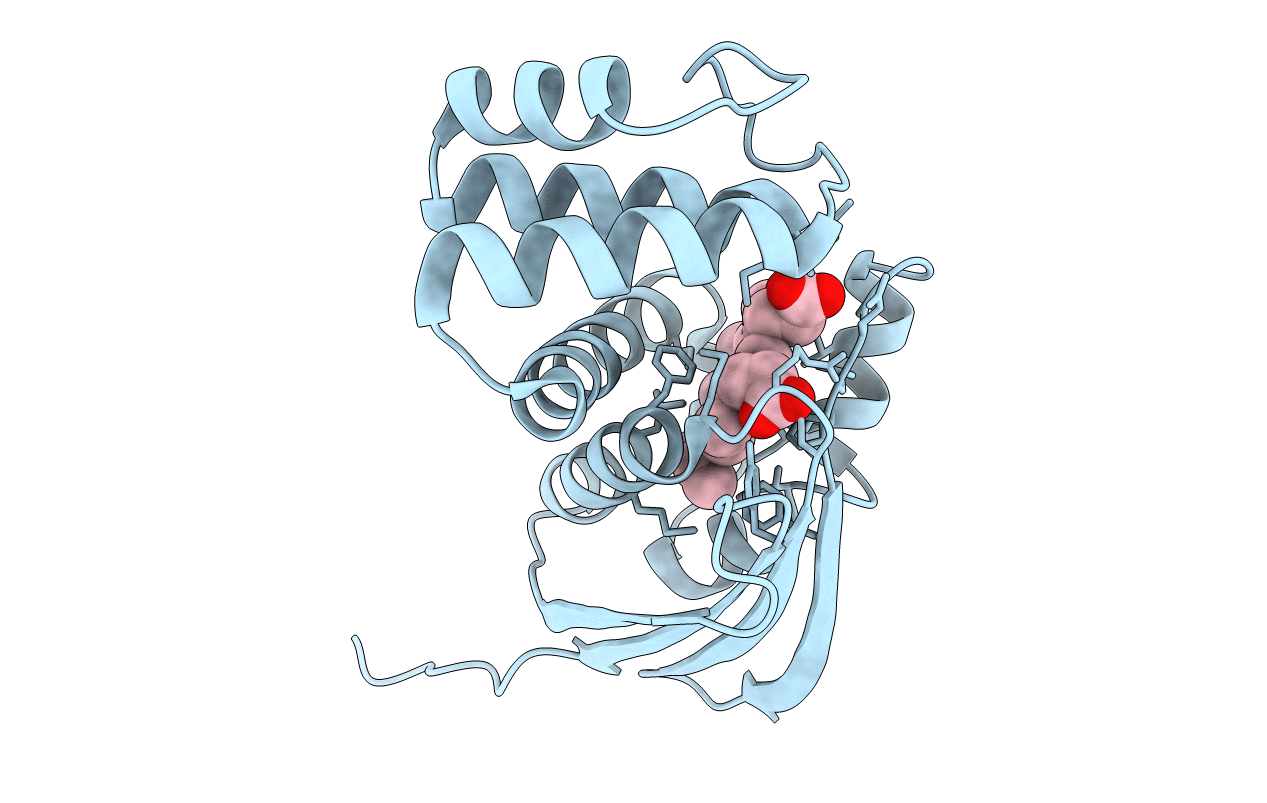 |
Crystal Structure Of Fe(Ii) Unliganded H-Nox Protein Mutant A71G From K. Algicida
Organism: Kordia algicida ot-1
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2018-05-30 Classification: SIGNALING PROTEIN Ligands: HEM |
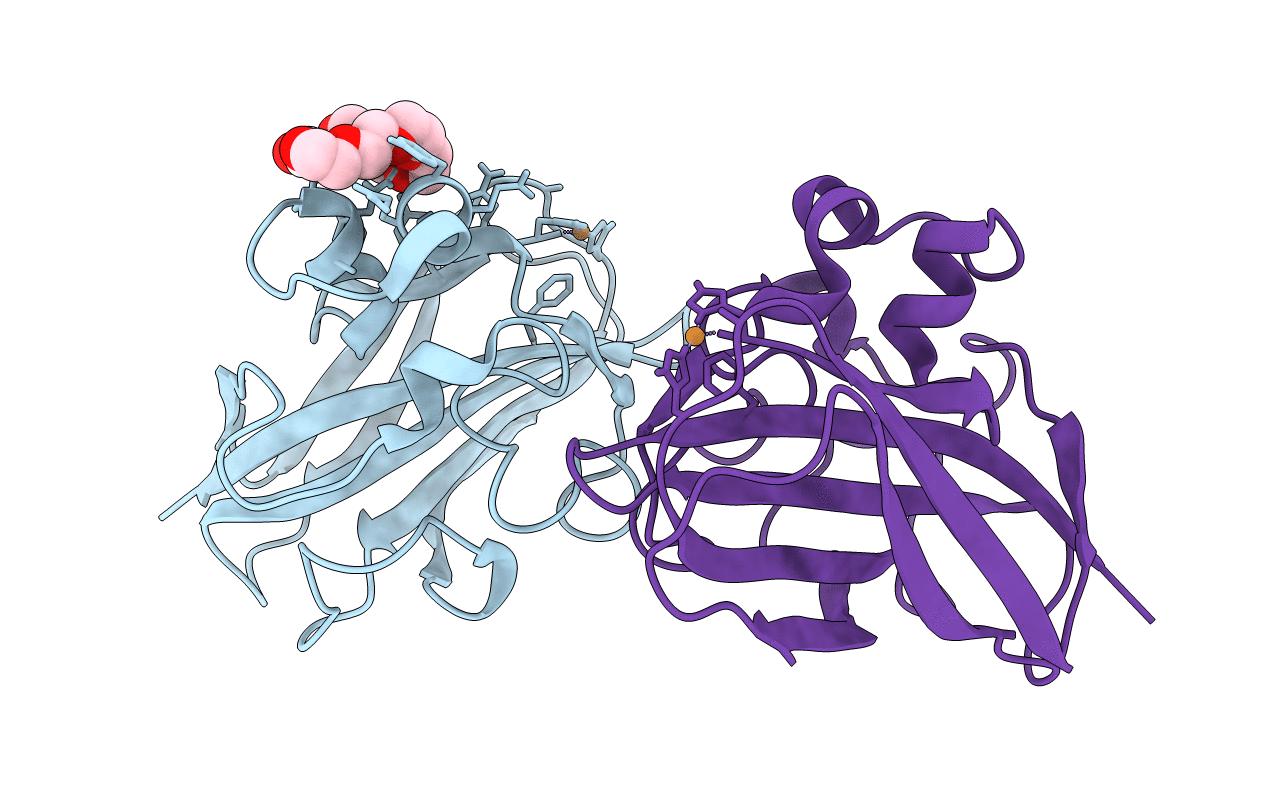 |
Organism: Listeria monocytogenes serotype 1/2a (strain 10403s)
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2017-08-09 Classification: CHITIN-BINDING PROTEIN Ligands: CU, P33 |
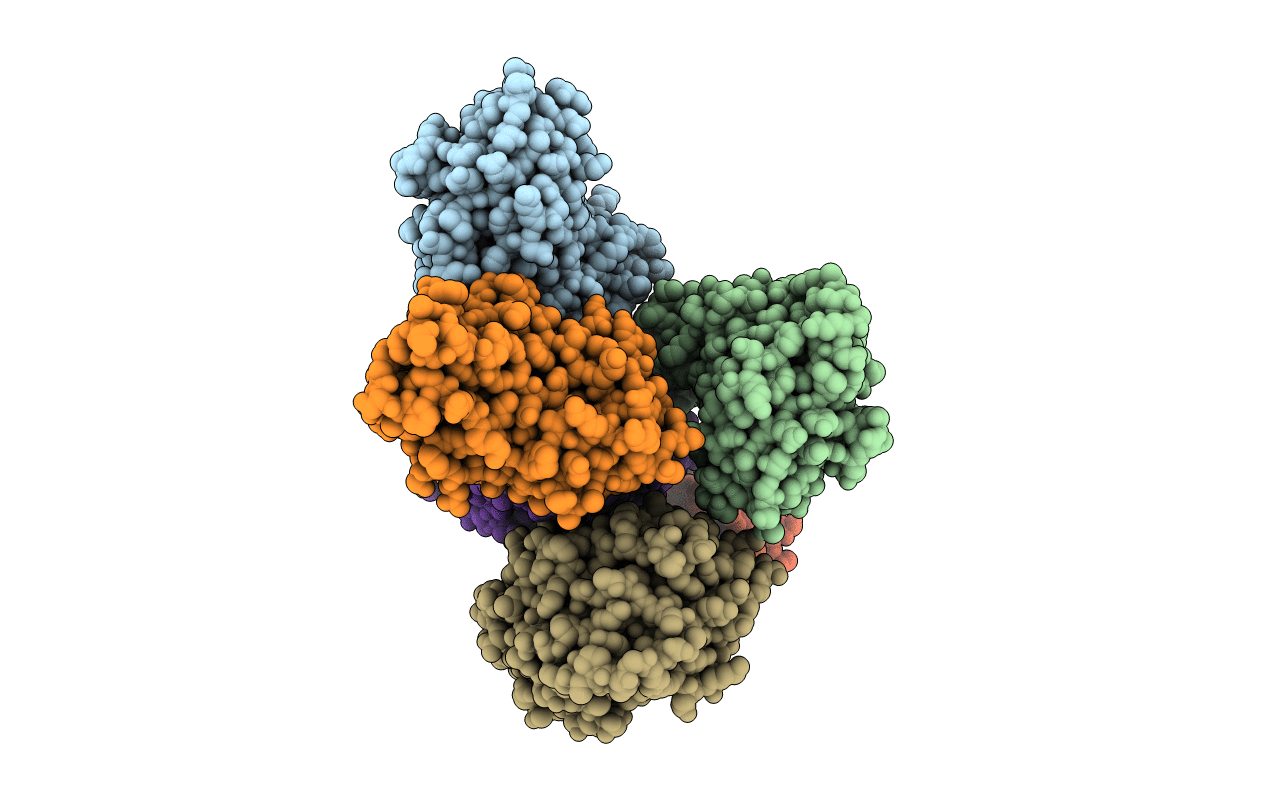 |
Crystal Structure Of A Cellulose-Active Polysaccharide Monooxygenase From M. Thermophila (Mtpmo3*)
Organism: Myceliophthora thermophila
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2017-03-15 Classification: Hydrolase/OXIDOREDUCTASE Ligands: CU |
 |
Organism: Caldanaerobacter subterraneus subsp. tengcongensis (strain dsm 15242 / jcm 11007 / nbrc 100824 / mb4)
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2016-07-06 Classification: SIGNALING PROTEIN Ligands: HEM |
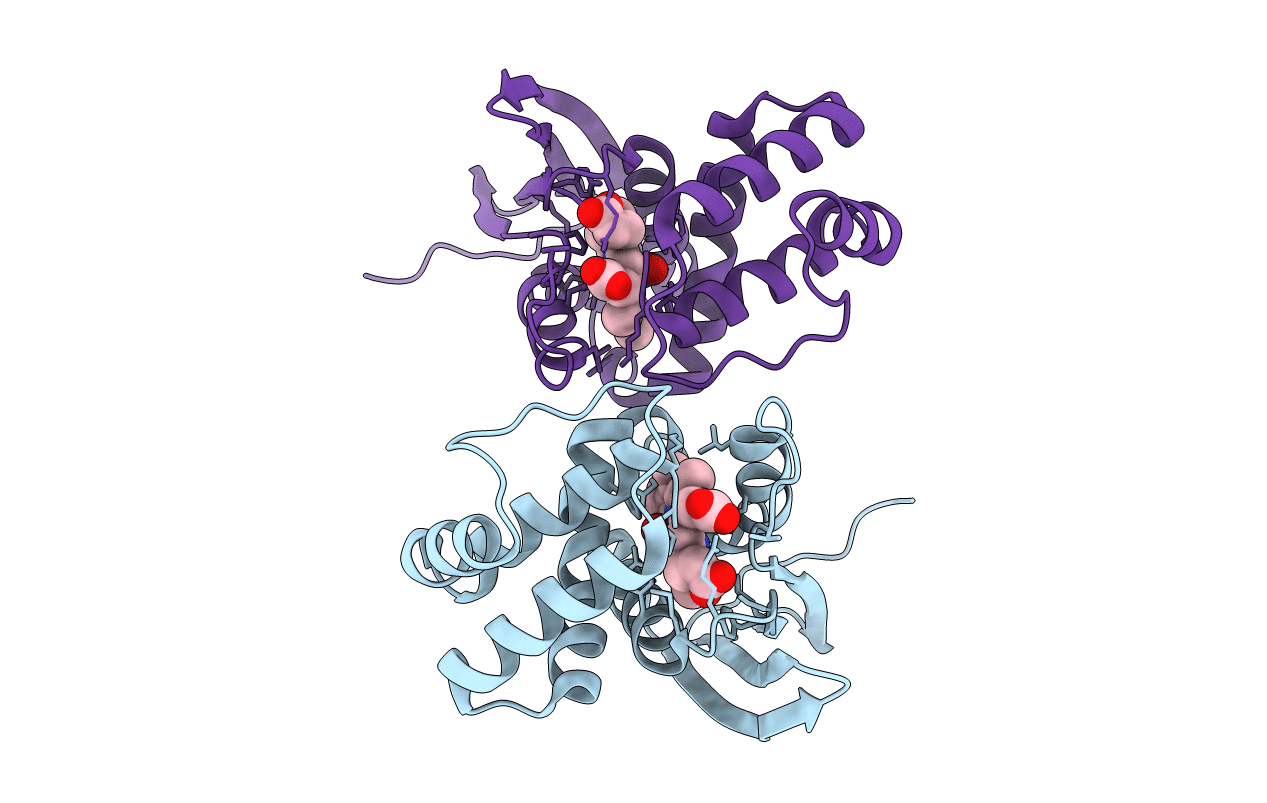 |
Organism: Caldanaerobacter subterraneus subsp. tengcongensis (strain dsm 15242 / jcm 11007 / nbrc 100824 / mb4)
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2016-07-06 Classification: SIGNALING PROTEIN Ligands: HEM, NO, IOD |
 |
Organism: Caldanaerobacter subterraneus subsp. tengcongensis (strain dsm 15242 / jcm 11007 / nbrc 100824 / mb4)
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2016-07-06 Classification: SIGNALING PROTEIN Ligands: HEM, CMO |
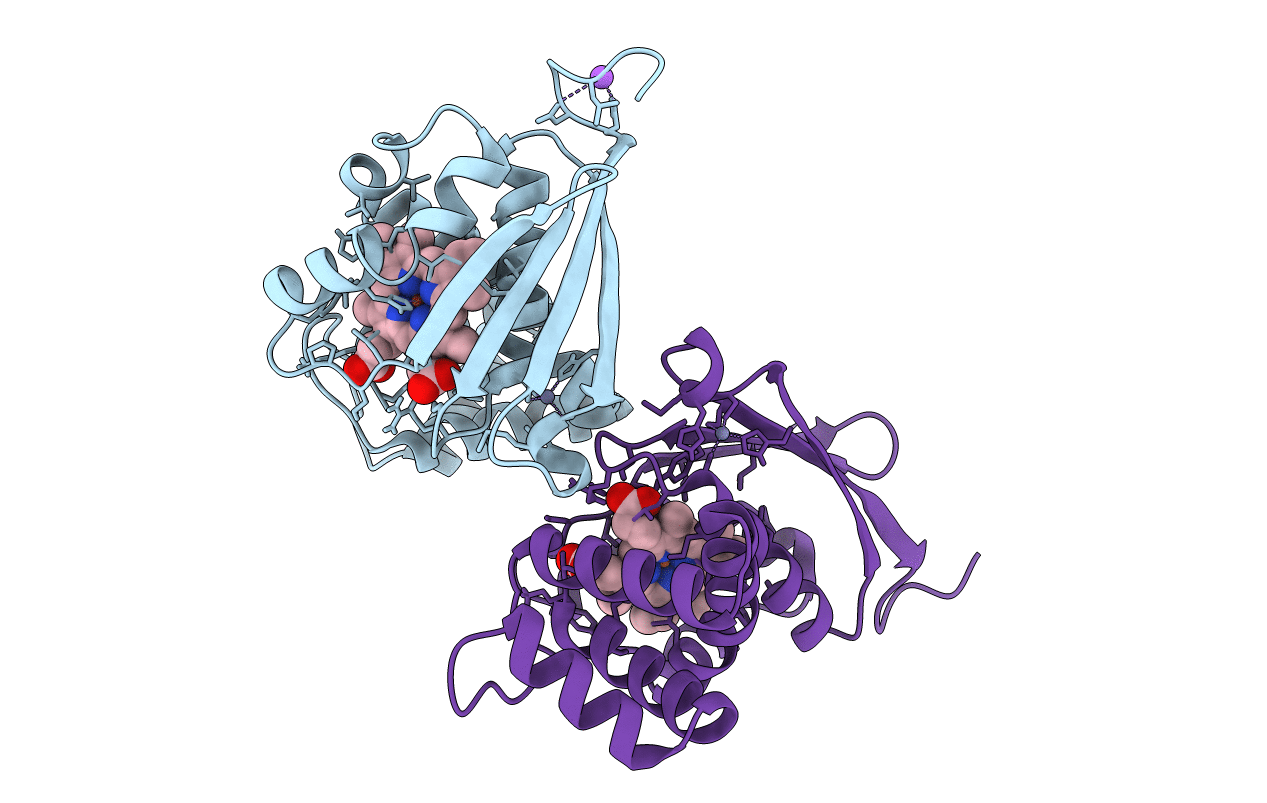 |
Crystal Structure Of An H-Nox Protein From S. Oneidensis In The Fe(Ii) Ligation State, Q154A/Q155A/K156A Mutant
Organism: Shewanella oneidensis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2014-10-01 Classification: SIGNALING PROTEIN Ligands: HEM, ZN, NA |
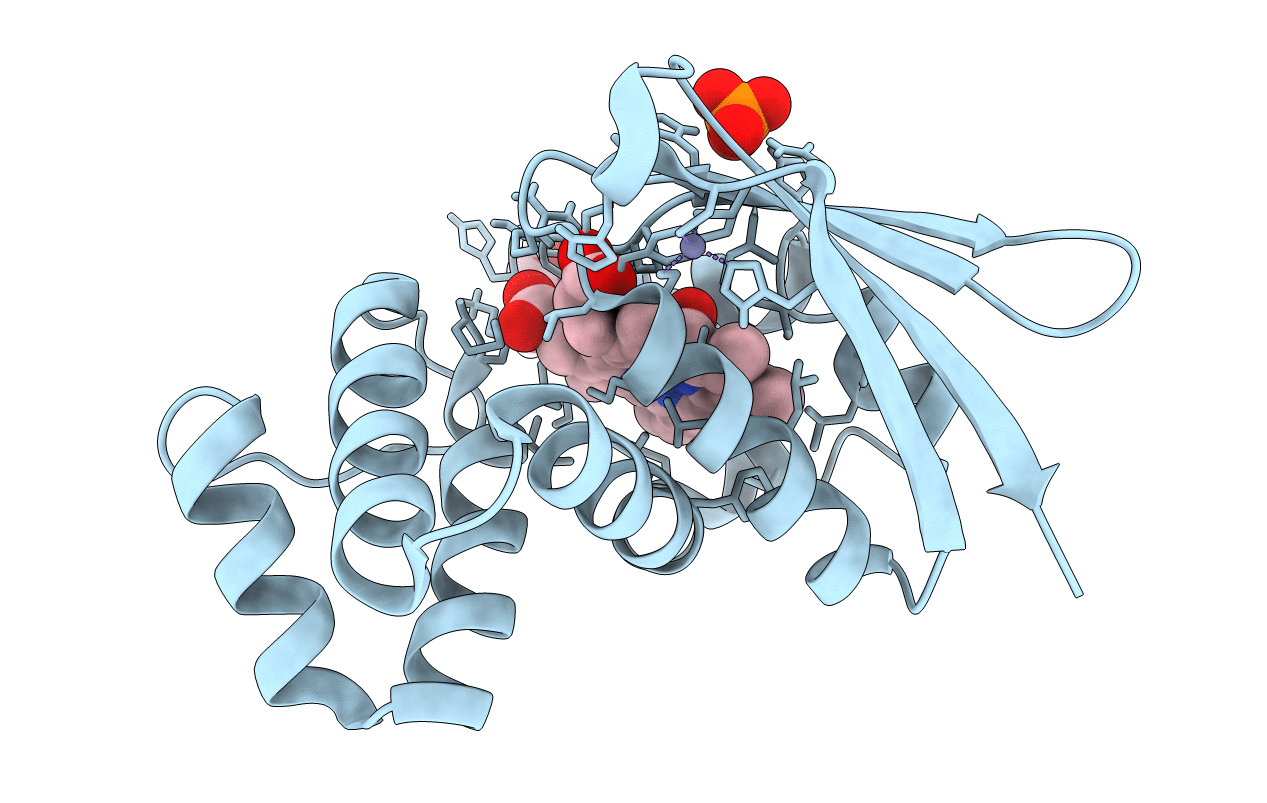 |
Crystal Structure Of An H-Nox Protein From S. Oneidensis In The Fe(Ii)No Ligation State
Organism: Shewanella oneidensis
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2014-10-01 Classification: SIGNALING PROTEIN Ligands: HEM, NO, ZN, PO4, GOL |
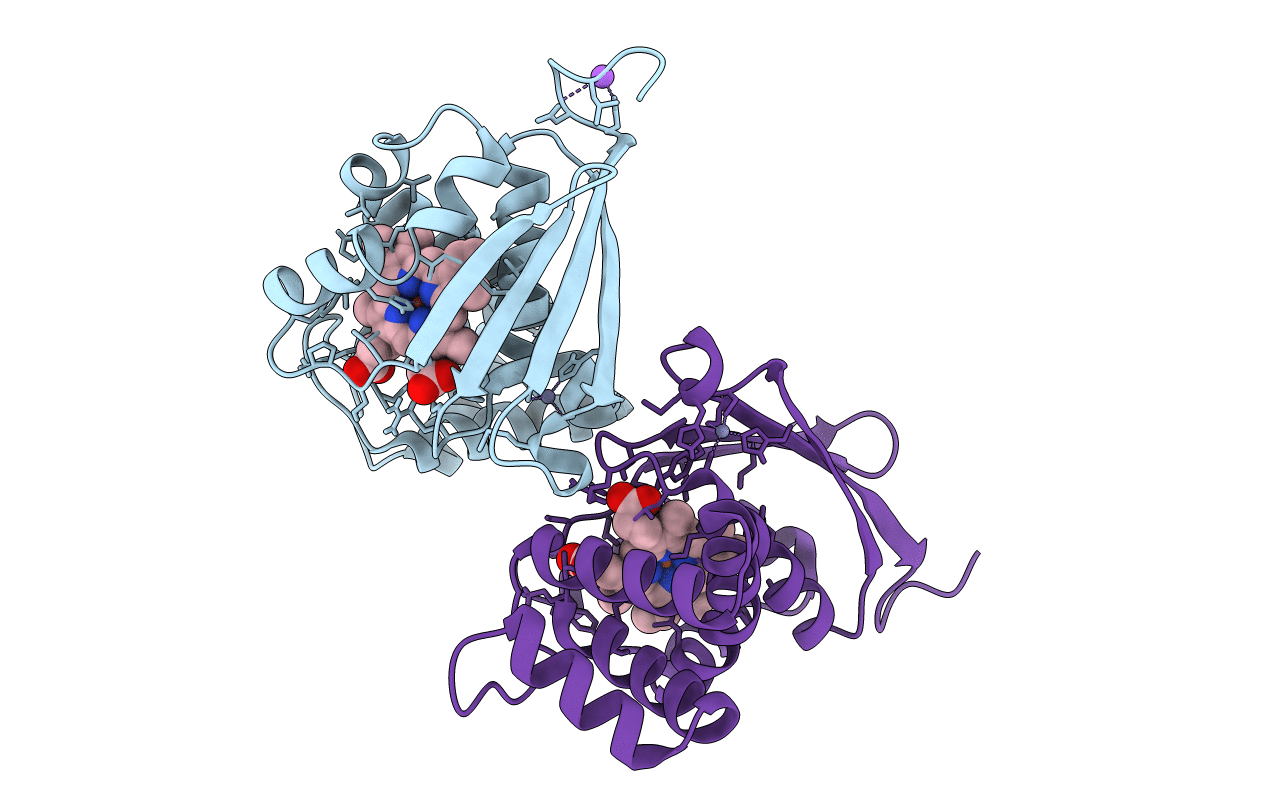 |
Crystal Structure Of An H-Nox Protein From S. Oneidensis In The Fe(Ii)Co Ligation State, Q154A/Q155A/K156A Mutant
Organism: Shewanella oneidensis
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2014-10-01 Classification: SIGNALING PROTEIN Ligands: HEM, ZN, CMO, NA |

