Search Count: 6
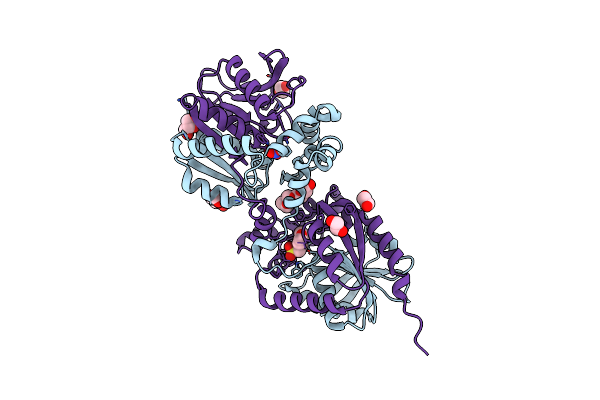 |
Crystal Structure Of Haloalkane Dehalogenase Variant Dhaa115 Domain-Swapped Dimer Type-1
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2021-01-27 Classification: HYDROLASE Ligands: EDO, MES, GOL, PG4, NO3 |
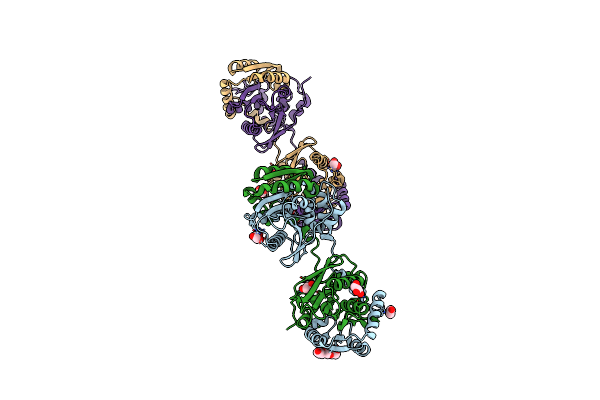 |
Crystal Structure Of Haloalkane Dehalogenase Variant Dhaa115 Domain-Swapped Dimer Type-2
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2021-01-27 Classification: HYDROLASE Ligands: CL, GOL, PG4, EDO |
 |
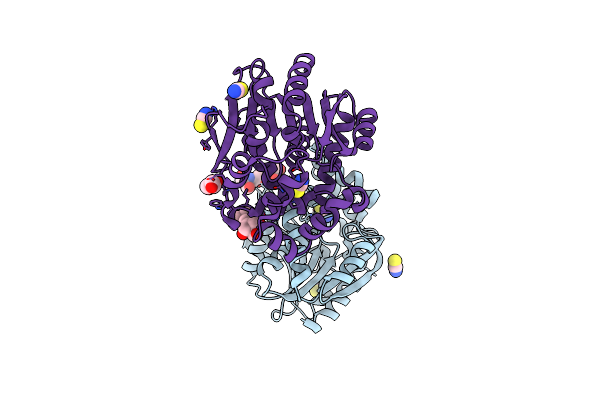
Crystal Structure Of Haloalkane Dehalogenase Variant Dhaa177 Domain-Swapped Dimer Type-3
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.54 Å Release Date: 2021-01-27 Classification: HYDROLASE Ligands: GOL, SO4 |
 |
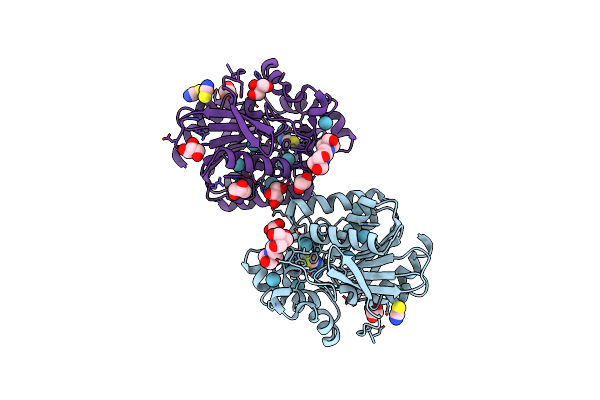
Organism: Rhodococcus rhodochrous
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2020-11-18 Classification: HYDROLASE Ligands: B3P, SCN, GOL |
 |
Structure Of Hyperstable Haloalkane Dehalogenase Variant Dhaa115 Prepared By The 'Soak-And-Freeze' Method Under 150 Bar Of Krypton Pressure
Organism: Rhodococcus rhodochrous
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2020-11-18 Classification: HYDROLASE Ligands: KR, SCN, B3P, GOL |
 |
Crystal Structure Of The Common Ancestor Of Haloalkane Dehalogenases And Renilla Luciferase (Anchld-Rluc)
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.39 Å Release Date: 2019-04-24 Classification: HYDROLASE Ligands: OXY, MPD, TRS |

