Search Count: 388
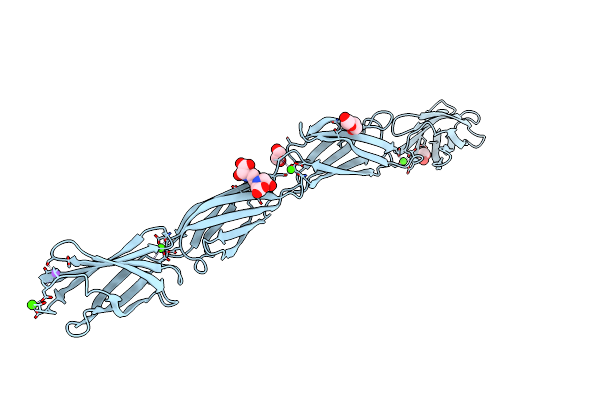 |
Crystal Structure Of Tetra-Tandem Repeat In Extending Region Of Large Adhesion Protein
Organism: Marinobacter hydrocarbonoclasticus (strain atcc 49840 / dsm 8798 / sp17)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-10-14 Classification: METAL BINDING PROTEIN Ligands: CA, CL, NA, EDT, PEG, EDO |
 |
Organism: Marinobacter hydrocarbonoclasticus atcc 49840
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2019-09-04 Classification: SUGAR BINDING PROTEIN Ligands: BGC, CA, NA |
 |
Wax Ester Synthase/Diacylglycerol Acyltransferase From Marinobacter Aquaeolei Vt8
Organism: Marinobacter hydrocarbonoclasticus (strain atcc 700491 / dsm 11845 / vt8)
Method: X-RAY DIFFRACTION Resolution:2.43 Å Release Date: 2018-06-13 Classification: TRANSFERASE Ligands: CL |
 |
X-Ray Crystal Structure Of Fatty Aldehyde Dehydrogenase Enzymes From Marinobacter Aquaeolei Vt8 Complexed With A Substrate
Organism: Marinobacter hydrocarbonoclasticus (strain atcc 700491 / dsm 11845 / vt8)
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2017-04-26 Classification: OXIDOREDUCTASE Ligands: EDO, 8YP, MPD, CL, PO4 |
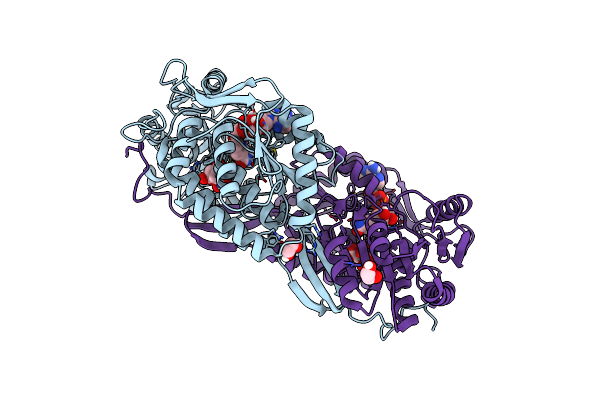 |
Fatty Aldehyde Dehydrogenase From Marinobacter Aquaeolei Vt8 And Cofactor Complex
Organism: Marinobacter hydrocarbonoclasticus (strain atcc 700491 / dsm 11845 / vt8)
Method: X-RAY DIFFRACTION Resolution:3.08 Å Release Date: 2017-04-26 Classification: OXIDOREDUCTASE Ligands: EDO, CIT, NAD |
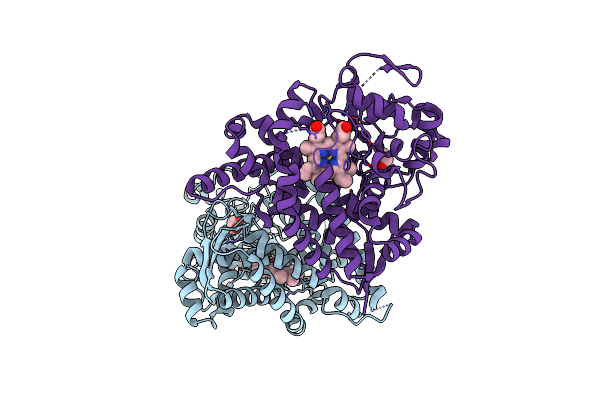 |
Organism: Marinobacter hydrocarbonoclasticus
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2017-01-18 Classification: OXIDOREDUCTASE Ligands: HEM, EDO |
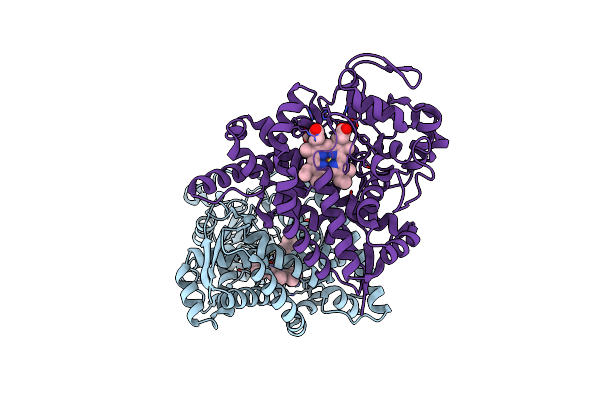 |
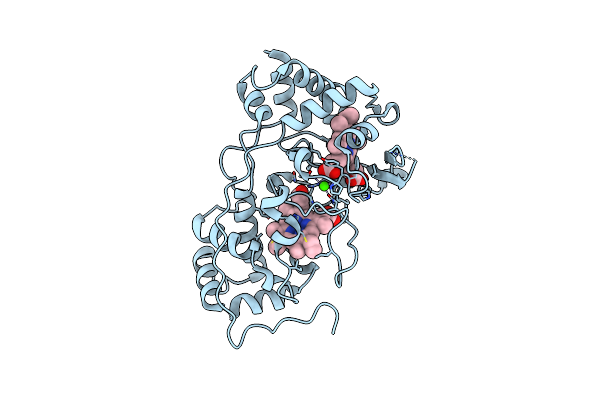
Structure Of Cyp153A From Marinobacter Aquaeolei In Complex With Hydroxydodecanoic Acid
Organism: Marinobacter hydrocarbonoclasticus
Method: X-RAY DIFFRACTION Resolution:2.22 Å Release Date: 2017-01-18 Classification: OXIDOREDUCTASE Ligands: HEM, 12H |
 |
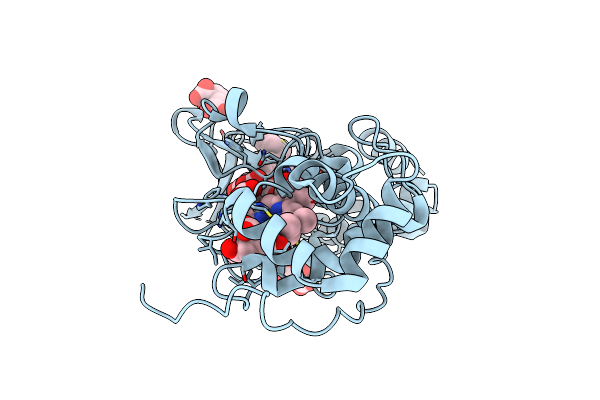
Organism: Marinobacter hydrocarbonoclasticus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2004-06-29 Classification: OXIDOREDUCTASE Ligands: CA, HEC |
 |
Organism: Marinobacter hydrocarbonoclasticus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2004-06-29 Classification: OXIDOREDUCTASE Ligands: HEC, CIT |
 |
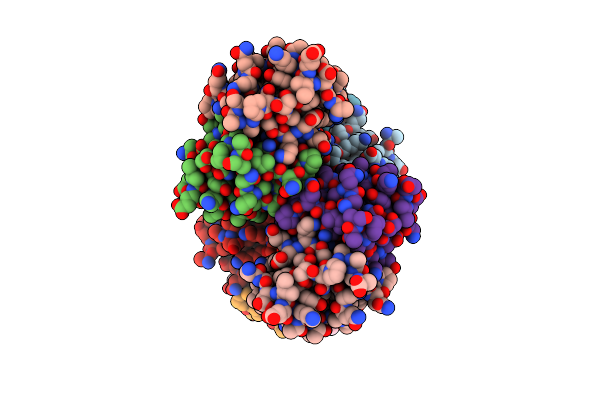
Di-Haemic Cytochrome C Peroxidase From Pseudomonas Nautica 617, Form In (Ph 4.0)
Organism: Marinobacter hydrocarbonoclasticus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2004-01-13 Classification: OXIDOREDUCTASE Ligands: HEC, CIT |
 |
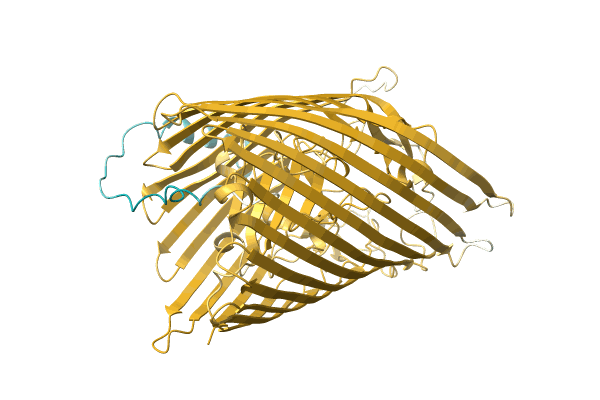
Organism: Marinobacter hydrocarbonoclasticus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 1999-07-22 Classification: ELECTRON TRANSPORT Ligands: HEC, GOL |
 |
Organism: Marinobacter hydrocarbonoclasticus
Method: Alphafold Release Date: Classification: NA Ligands: NA |
 |
Organism: Marinobacter hydrocarbonoclasticus
Method: Alphafold Release Date: Classification: NA Ligands: NA |
 |
Organism: Marinobacter hydrocarbonoclasticus
Method: Alphafold Release Date: Classification: NA Ligands: NA |
 |
NA
Organism: Marinobacter hydrocarbonoclasticus (strain ATCC 700491 / DSM 11845 / VT8)
Method: Alphafold Release Date: Classification: NA Ligands: NA |
 |
NA
Organism: Marinobacter hydrocarbonoclasticus (strain ATCC 700491 / DSM 11845 / VT8)
Method: Alphafold Release Date: Classification: NA Ligands: NA |
 |
NA
Organism: Marinobacter hydrocarbonoclasticus (strain ATCC 700491 / DSM 11845 / VT8)
Method: Alphafold Release Date: Classification: NA Ligands: NA |
 |
NA
Organism: Marinobacter hydrocarbonoclasticus (strain ATCC 700491 / DSM 11845 / VT8)
Method: Alphafold Release Date: Classification: NA Ligands: NA |
 |
NA
Organism: Marinobacter hydrocarbonoclasticus (strain ATCC 700491 / DSM 11845 / VT8)
Method: Alphafold Release Date: Classification: NA Ligands: NA |
 |
NA
Organism: Marinobacter hydrocarbonoclasticus (strain ATCC 700491 / DSM 11845 / VT8)
Method: Alphafold Release Date: Classification: NA Ligands: NA |

