Search Count: 46
 |
Crystal Structure Of Iron-Bound Human Ado C18S/C239S Variant Soaked In Hydralazine At 2.39 Angstrom Resolution
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-09-24 Classification: OXIDOREDUCTASE Ligands: FE2, HLZ, GOL |
 |
Crystal Structure Of Cobalt-Bound Human Ado C18S/C239S Variant In Complex With Hydralazine At 1.89 Angstrom Resolution
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-09-10 Classification: OXIDOREDUCTASE Ligands: CO, HLZ, GOL, SO4 |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-06-18 Classification: TRANSPORT PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-06-18 Classification: TRANSPORT PROTEIN Ligands: R75 |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-06-18 Classification: TRANSPORT PROTEIN Ligands: A1AIL |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-06-18 Classification: TRANSPORT PROTEIN Ligands: A1A45 |
 |
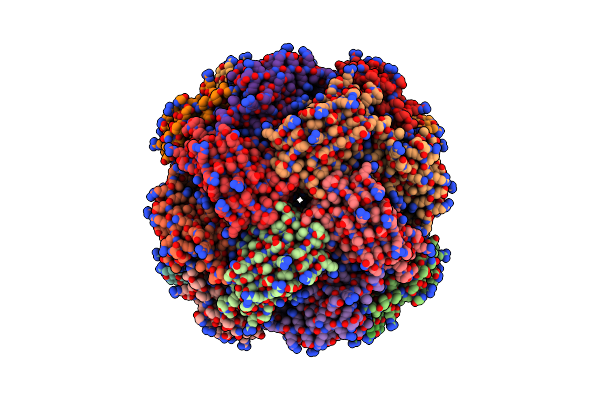
Imidazole Glycerol Phosphate Dehydratase From Mycobacterium Tuberculosis, Apo Structure
Organism: Mycobacterium tuberculosis
Method: ELECTRON MICROSCOPY Release Date: 2025-06-18 Classification: LYASE Ligands: MN |
 |
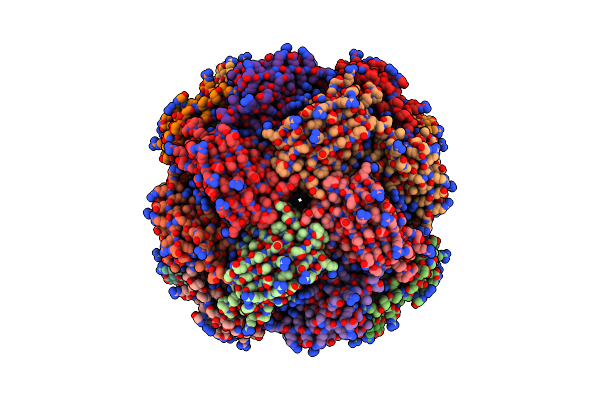
Imidazole Glycerol Phosphate Dehydratase From Mycobacterium Tuberculosis, In Complex With Aminotriazole
Organism: Mycobacterium tuberculosis
Method: ELECTRON MICROSCOPY Release Date: 2025-06-18 Classification: LYASE Ligands: MN, 3TR |
 |
Imidazole Glycerol Phosphate Dehydratase From Mycobacterium Tuberculosis, Apo Structure
Organism: Mycobacterium tuberculosis
Method: ELECTRON MICROSCOPY Release Date: 2025-06-18 Classification: LYASE Ligands: MN |
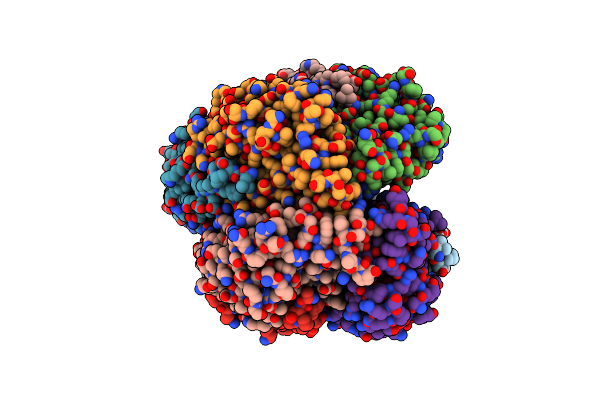 |
Organism: Hypocrea virens (strain gv29-8 / fgsc 10586)
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2025-04-23 Classification: DNA BINDING PROTEIN |
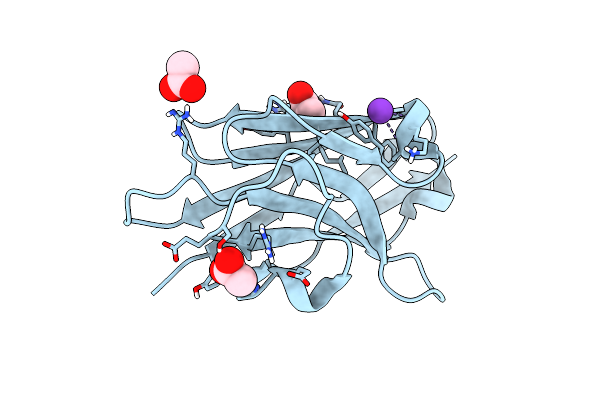 |
Organism: Camelus dromedarius
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2024-07-10 Classification: IMMUNE SYSTEM Ligands: ACT, K |
 |
Organism: Arabidopsis thaliana, Saccharomyces cerevisiae s288c , Saccharomyces cerevisiae, Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2024-02-14 Classification: TRANSFERASE/INHIBITOR Ligands: PLP, Z1T |
 |
Organism: Arabidopsis thaliana, Saccharomyces cerevisiae s288c, Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2024-02-14 Classification: TRANSFERASE/INHIBITOR |
 |
Organism: Arabidopsis thaliana, Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2024-02-14 Classification: TRANSFERASE/INHIBITOR Ligands: PLP, Z1T |
 |
Crystal Structure Of N-Terminal Domain Of Nucleocapsid Protein Of Sars-Cov-2
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-11-22 Classification: RNA BINDING PROTEIN Ligands: GOL |
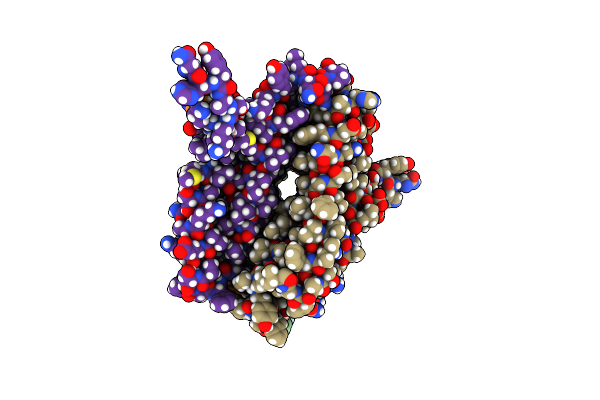 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-05-17 Classification: MEMBRANE PROTEIN |
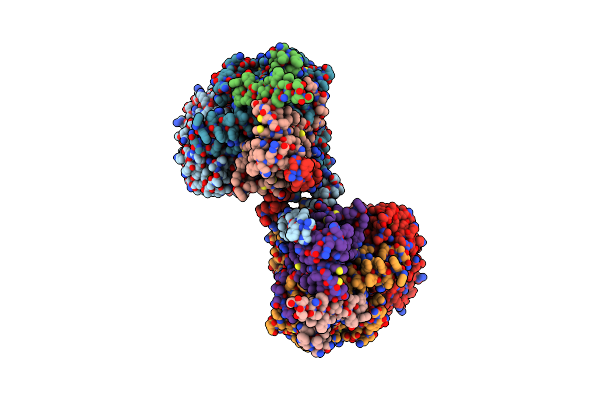 |
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2023-04-05 Classification: TRANSFERASE/INHIBITOR Ligands: Z1T |
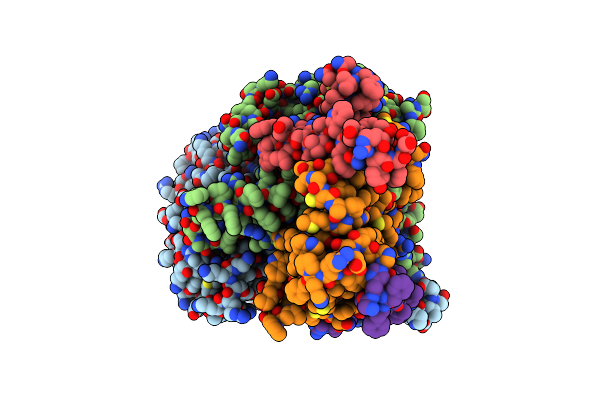 |
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2023-04-05 Classification: TRANSFERASE/INHIBITOR Ligands: PLP, Z1T |
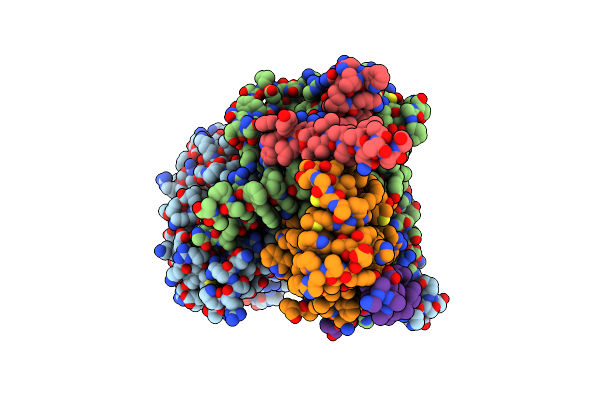 |
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2023-04-05 Classification: TRANSFERASE/INHIBITOR Ligands: PLP |
 |
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2023-04-05 Classification: TRANSFERASE/INHIBITOR Ligands: PLS, Z1T |

