Search Count: 76
 |
Crystal Structure Of Renilla Luciferase Rluc8-Gfp Bret Complex At Ph 9.0 (Space Group P32)
Organism: Renilla reniformis
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: HYDROLASE Ligands: CEI |
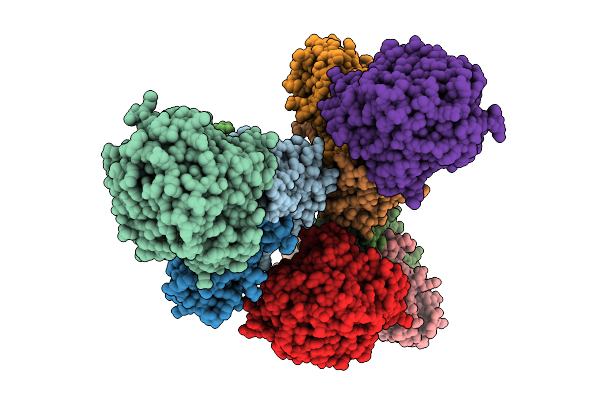 |
Organism: Renilla reniformis
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: HYDROLASE Ligands: CEI |
 |
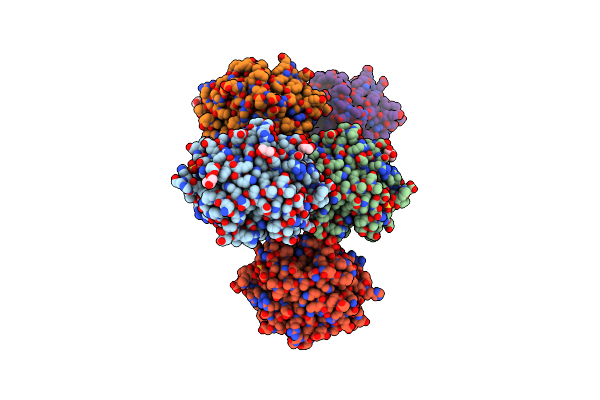
Crystal Structure Of Renilla Reniformis Luciferase Rluc8-Gfp Bret Complex At Ph 6.0
Organism: Renilla reniformis
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: HYDROLASE Ligands: CEI |
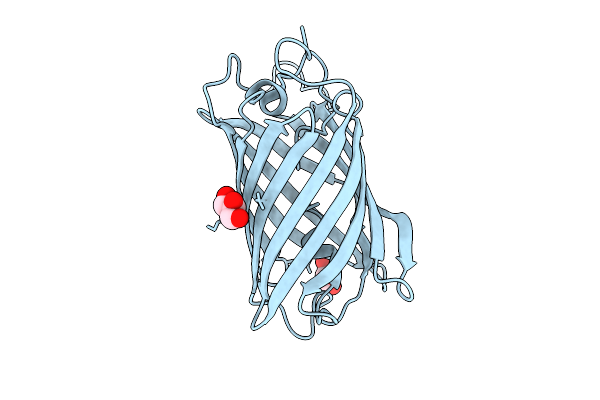 |
Crystal Structure Of Full-Lenght Renilla Reniformis Green Fluorescent Protein (Gfp)
Organism: Renilla reniformis
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: FLUORESCENT PROTEIN Ligands: GOL |
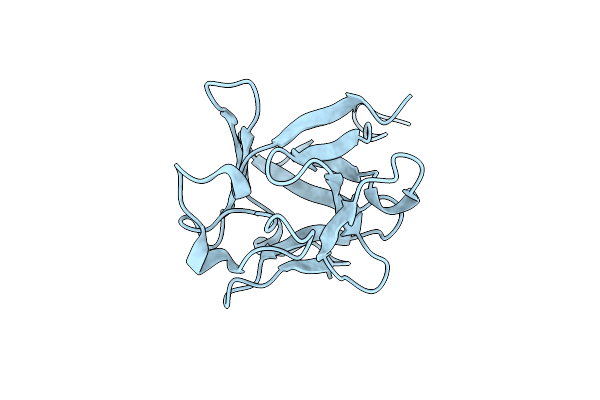 |
Crystal Structure Of Fgf2-Stab, A Stable Variant Of Human Fibroblast Growth Factor 2
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.31 Å Release Date: 2024-03-06 Classification: CYTOKINE |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2024-01-17 Classification: HYDROLASE Ligands: CL, SO4, PG4, GOL, TRS |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.31 Å Release Date: 2024-01-17 Classification: HYDROLASE Ligands: MG, CL |
 |
Cryo-Em Map Of The Focused Refinement Of The Subfamily Iii Haloalkane Dehalogenase From Haloferax Mediterranei Dimer Forming Hexameric Assembly.
Organism: Haloferax mediterranei
Method: ELECTRON MICROSCOPY Release Date: 2023-10-11 Classification: HYDROLASE |
 |
Nanoluc-D9R/H57A/K89R Mutant Complexed With Azacoelenterazine Bound In Intra-Barrel Catalytic Site
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2023-09-27 Classification: LUMINESCENT PROTEIN Ligands: NSW |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2023-08-30 Classification: LIPID BINDING PROTEIN |
 |
Structure Of Haloalkane Dehalogenase Dmmara From Mycobacterium Marinum At Ph 6.5
Organism: Mycobacterium marinum
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2023-08-30 Classification: HYDROLASE Ligands: GOL, FMT, TRS, B3P |
 |
Structure Of Haloalkane Dehalogenase Dmmara From Mycobacterium Marinum At Ph 5.5
Organism: Mycobacterium marinum
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2023-08-30 Classification: HYDROLASE Ligands: GOL, FMT, ACT |
 |
X-Ray Structure Of The Crystallization-Prone Form Of Subfamily Iii Haloalkane Dehalogenase Dhmea From Haloferax Mediterranei
Organism: Haloferax mediterranei
Method: X-RAY DIFFRACTION Resolution:3.31 Å Release Date: 2023-08-30 Classification: UNKNOWN FUNCTION Ligands: CL |
 |
Organism: Oplophorus gracilirostris
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2023-08-23 Classification: LUMINESCENT PROTEIN Ligands: GOL, PG4, OXY, CL, PGE, NT0 |
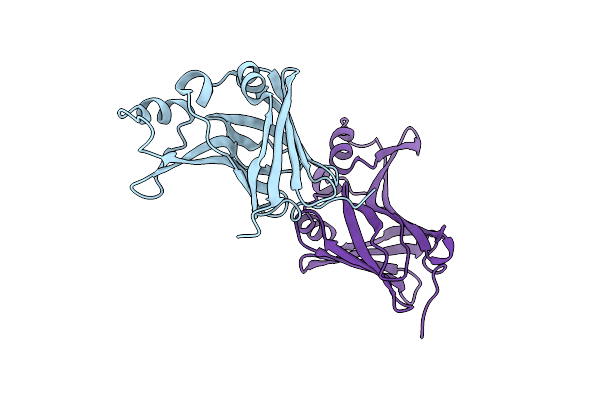 |
Organism: Oplophorus gracilirostris
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2023-08-23 Classification: LUMINESCENT PROTEIN |
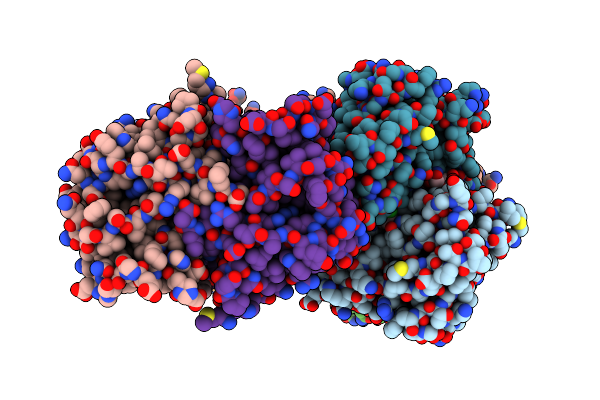 |
Organism: Oplophorus gracilirostris
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2023-08-23 Classification: LUMINESCENT PROTEIN Ligands: CEI, CL, PG4 |
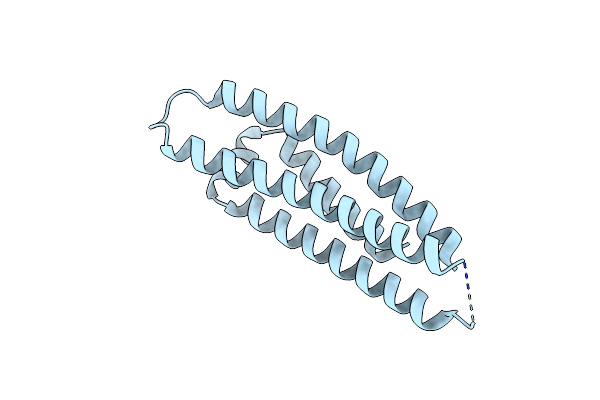 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2023-08-02 Classification: LIPID BINDING PROTEIN |
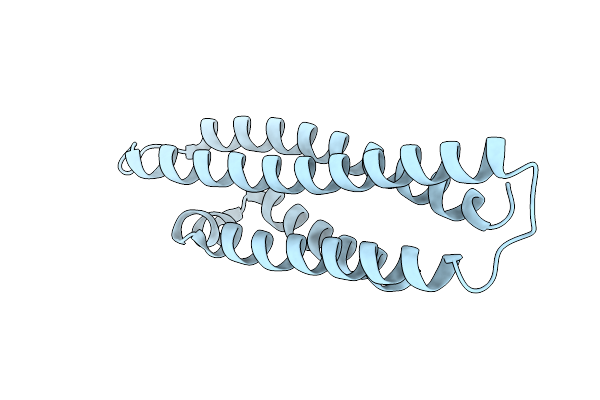 |
Organism: Escherichia coli, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2023-07-19 Classification: LIPID TRANSPORT |
 |
Organism: Escherichia coli, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2023-07-19 Classification: LIPID TRANSPORT Ligands: GOL |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2022-12-07 Classification: OXIDOREDUCTASE Ligands: CEI |

