Search Count: 56
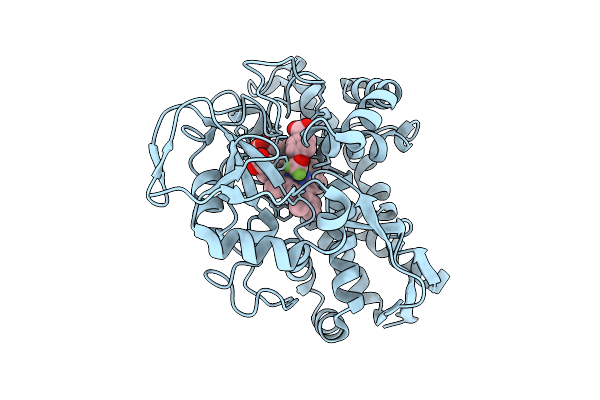 |
The Crystal Structure Of Wild-Type Cyp199A4 Bound To 4-Trifluoromethoxybenzoic Acid
Organism: Rhodopseudomonas palustris haa2
Method: X-RAY DIFFRACTION Release Date: 2025-08-13 Classification: OXIDOREDUCTASE Ligands: M4F, HEM, CL |
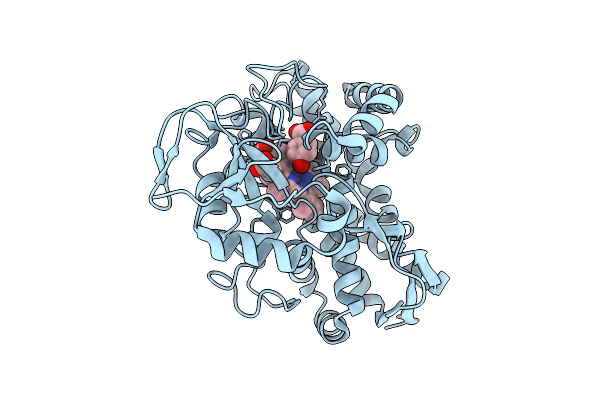 |
Organism: Rhodopseudomonas palustris haa2
Method: X-RAY DIFFRACTION Release Date: 2025-08-13 Classification: OXIDOREDUCTASE Ligands: PHB, HEM, CL |
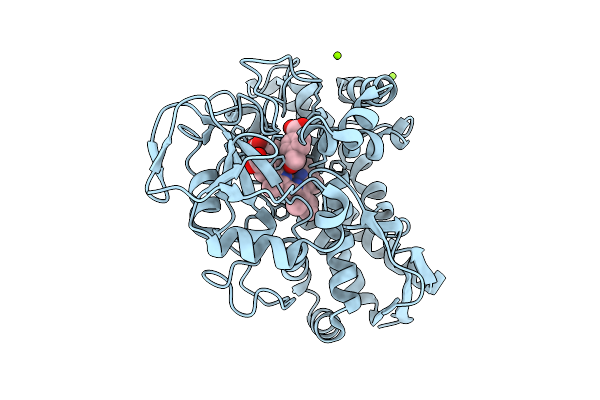 |
The Crystal Structure Of Wild-Type Cyp199A4 Bound To 4-(Hydroxymethyl)Benzoic Acid
Organism: Rhodopseudomonas palustris haa2
Method: X-RAY DIFFRACTION Release Date: 2025-08-13 Classification: OXIDOREDUCTASE Ligands: E5X, HEM, CL, MG |
 |
Crystal Structure Of Artificial Enzyme Lmrr_Paf Variant Rmh In Crystal Form 1
Organism: Lactococcus cremoris subsp. cremoris mg1363
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2025-02-26 Classification: DNA BINDING PROTEIN |
 |
Crystal Structure Of Artificial Enzyme Lmrr_Paf Variant Rmh In Crystal Form 2
Organism: Lactococcus cremoris subsp. cremoris mg1363
Method: X-RAY DIFFRACTION Resolution:2.24 Å Release Date: 2025-02-26 Classification: DNA BINDING PROTEIN |
 |
Organism: Lactococcus cremoris subsp. cremoris mg1363
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2025-02-26 Classification: DNA BINDING PROTEIN |
 |
X-Ray Structure Of Lysozyme Obtained Upon Reaction With [Vivo(8-Hq)2] In Ethylene Glycol
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Release Date: 2025-02-12 Classification: HYDROLASE |
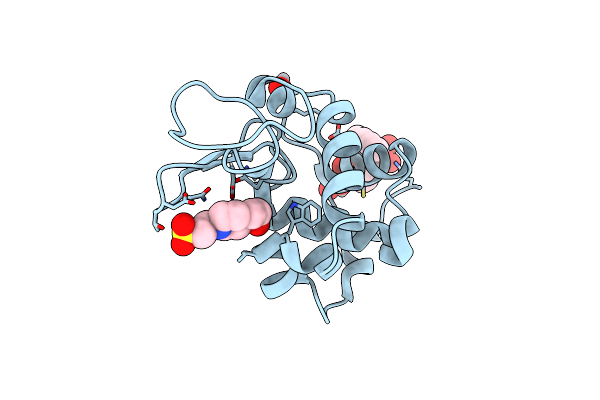 |
X-Ray Structure Of Lysozyme Obtained Upon Reaction With [Vivo(8-Hq)2] In Sodium Formate
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Release Date: 2025-02-12 Classification: HYDROLASE Ligands: EPE, A1H23, VVO, VVB |
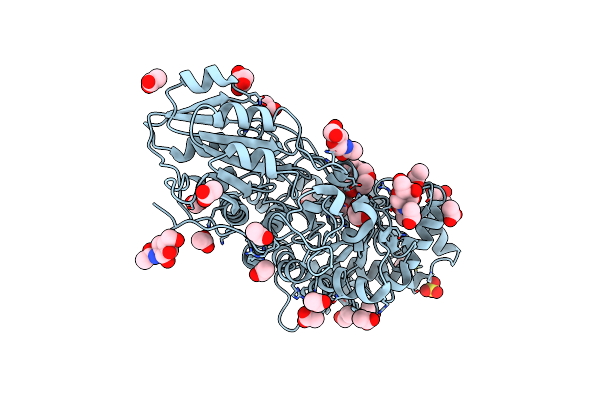 |
Organism: Hordeum vulgare
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2024-12-25 Classification: HYDROLASE Ligands: NAG, 1PE, GOL, ACT, SO4 |
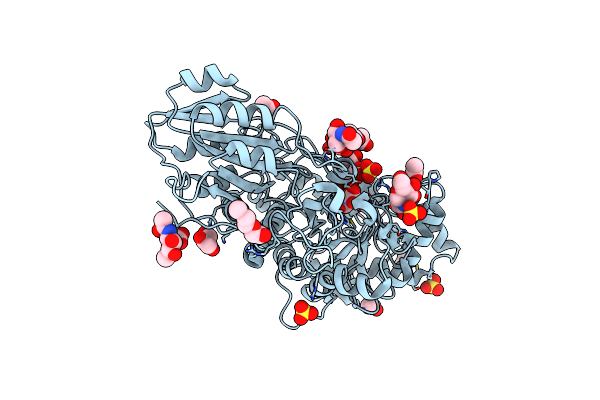 |
Crystal Structure Of Barley Exohydrolase Isoform Exoi E220A Mutant In Complex With Beta-D-Glucopyranose.
Organism: Hordeum vulgare
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2024-12-25 Classification: HYDROLASE Ligands: NAG, ACT, GOL, BGC, 1PE, SO4 |
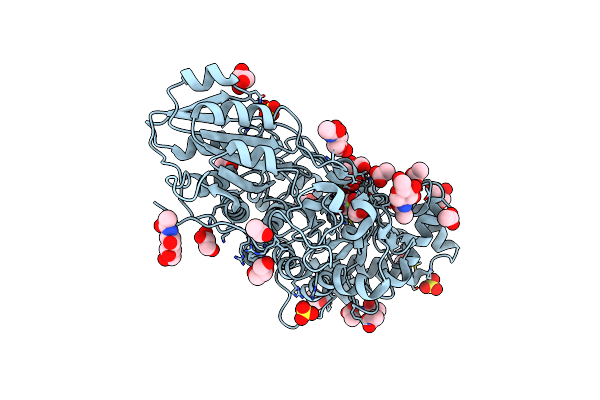 |
Crystal Structure Of Barley Exohydrolase Isoform Exoi E220A Mutant In Complex With 2-Deoxy-2-Fluoro-D-Glucopyranosides
Organism: Hordeum vulgare
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2024-12-25 Classification: HYDROLASE Ligands: NAG, G2F, SHG, GOL, ACT, SO4 |
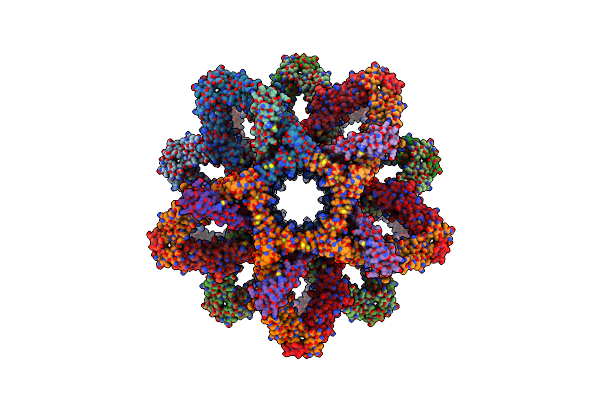 |
3D Reconstruction Of The Cylindrical Assembly Of Dnaja2 Delta G/F By Imposing D5 Symmetry
Organism: Saccharomyces cerevisiae s288c, Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:6.90 Å Release Date: 2023-07-26 Classification: CHAPERONE Ligands: ZN |
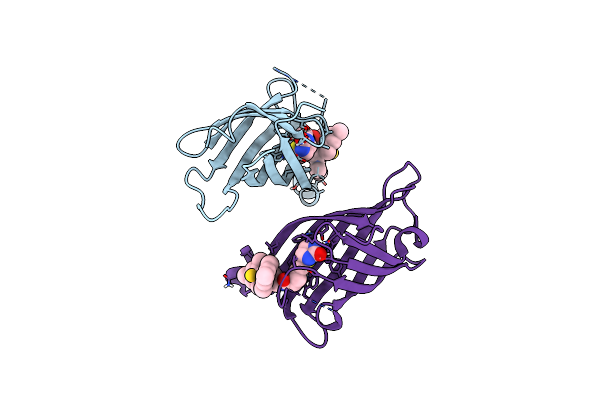 |
Organism: Streptomyces avidinii, Mycobacterium bovis (strain atcc baa-935 / af2122/97)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2021-07-14 Classification: METAL BINDING PROTEIN Ligands: UFW |
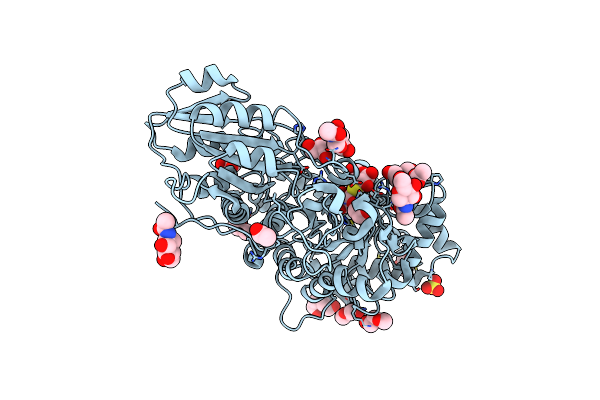 |
Crystal Structure Of Barley Exohydrolasei W434A Mutant In Complex With 4'-Nitrophenyl Thiolaminaritrioside
Organism: Hordeum vulgare subsp. vulgare
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-09-30 Classification: HYDROLASE Ligands: NAG, LAM, ACT, GOL, SO4, 1PE |
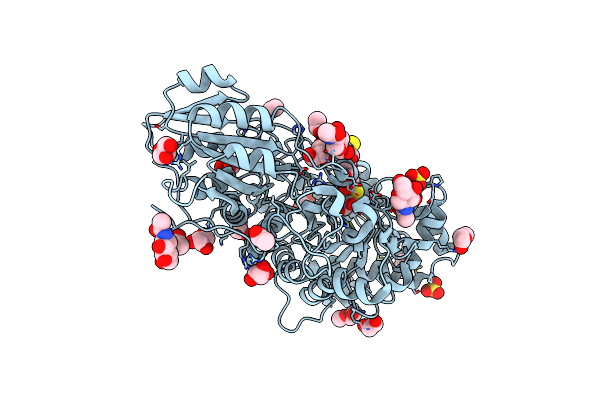 |
Crystal Structure Of Barley Exohydrolasei W434A Mutant In Complex With 4I,4Iii,4V-S-Trithiocellohexaose
Organism: Hordeum vulgare subsp. vulgare
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-09-30 Classification: HYDROLASE Ligands: NAG, GOL, SO4, 1PE, ACT |
 |
Crystal Structure Of Barley Exohydrolasei W434F Mutant In Complex With Methyl 6-Thio-Beta-Gentiobioside
Organism: Hordeum vulgare subsp. vulgare
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2020-09-30 Classification: HYDROLASE Ligands: GOL, 1PE, SO4, ACT |
 |
Crystal Structure Of Barley Exohydrolasei W434F In Complex With 4'-Nitrophenyl Thiolaminaribioside
Organism: Hordeum vulgare subsp. vulgare
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2020-09-30 Classification: HYDROLASE Ligands: 6BV, 1PE, GOL, SO4, EPE, ACT |
 |
Crystal Structure Of Barley Exohydrolasei W434A Mutant In Complex With Glucose.
Organism: Hordeum vulgare subsp. vulgare
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2020-09-02 Classification: HYDROLASE Ligands: BGC, NAG, ACT, GOL, SO4 |
 |
Crystal Structure Of Barley Exohydrolasei Wildtype In Complex With 4I,4Iii,4V-S-Trithiocellohexaose
Organism: Hordeum vulgare subsp. vulgare
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2020-08-19 Classification: HYDROLASE Ligands: NAG, 1PE, GOL, SO4 |
 |
Crystal Structure Of Barley Exohydrolasei Wildtype In Complex With 4'-Nitrophenyl Thiolaminaribioside
Organism: Hordeum vulgare subsp. vulgare
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-08-19 Classification: HYDROLASE Ligands: NAG, BV6, 1PE, SO4, GOL |

