Search Count: 17
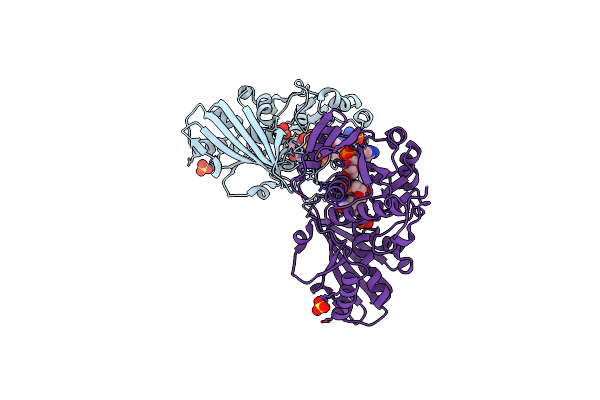 |
Crystal Structure Of Photosynthetic Glyceraldehyde-3-Phosphate Dehydrogenase From Chlamydomonas Reinhardtii (Crgapa) Complexed With Nadp+
Organism: Chlamydomonas reinhardtii
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2022-07-20 Classification: OXIDOREDUCTASE Ligands: NDP, SO4 |
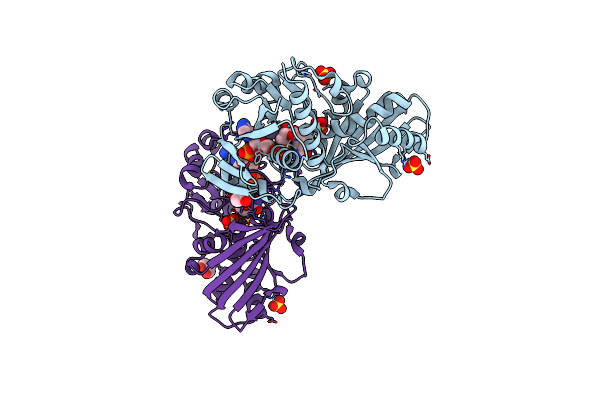 |
Crystal Structure Of Photosynthetic Glyceraldehyde-3-Phosphate Dehydrogenase From Chlamydomonas Reinhardtii (Crgapa) Complexed With Nadp+ And The Oxidated Catalytic Cysteine
Organism: Chlamydomonas reinhardtii
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2022-07-20 Classification: OXIDOREDUCTASE Ligands: NDP, SO4, EDO, GOL |
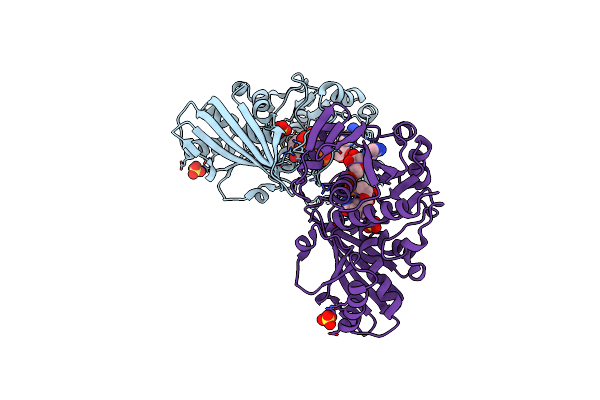 |
Crystal Structure Of Photosynthetic Glyceraldehyde-3-Phosphate Dehydrogenase From Chlamydomonas Reinhardtii (Crgapa) Complexed With Nad+
Organism: Chlamydomonas reinhardtii
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2022-07-20 Classification: OXIDOREDUCTASE Ligands: NAD, SO4 |
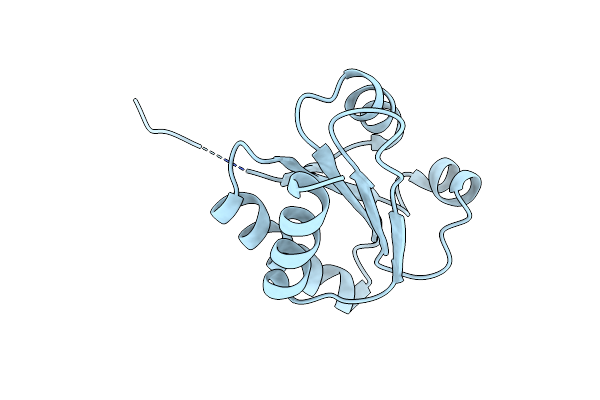 |
Crystal Structure Of Chloroplastic Thioredoxin Z Defines A Novel Type-Specific Target Recognition
Organism: Chlamydomonas reinhardtii
Method: X-RAY DIFFRACTION Resolution:2.44 Å Release Date: 2021-05-19 Classification: ISOMERASE Ligands: HOH |
 |
Crystal Structure Of Nitrosoglutathione Reductase (Gsnor) From Chlamydomonas Reinhardtii
Organism: Chlamydomonas reinhardtii
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-12-30 Classification: OXIDOREDUCTASE Ligands: ZN, PEG, CL |
 |
Crystal Structure Of Nitrosoglutathione Reductase From Chlamydomonas Reinhardtii In Complex With Nad+
Organism: Chlamydomonas reinhardtii
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2020-12-30 Classification: OXIDOREDUCTASE Ligands: ZN, NAD, PEG, CL, MG |
 |
Crystal Structure Of S-Nitrosylated Nitrosoglutathione Reductase(Gsnor)From Chlamydomonas Reinhardtii, In Complex With Nad+
Organism: Chlamydomonas reinhardtii
Method: X-RAY DIFFRACTION Release Date: 2020-12-30 Classification: OXIDOREDUCTASE Ligands: ZN, NAD, CL |
 |
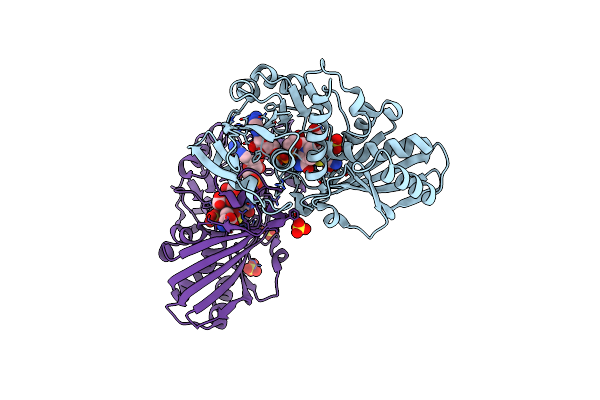
Crystal Structure Of Atgapc1 With The Catalytic Cys149 Irreversibly Oxidized By H2O2 Treatment
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2019-12-04 Classification: OXIDOREDUCTASE Ligands: NAD, SO4 |
 |
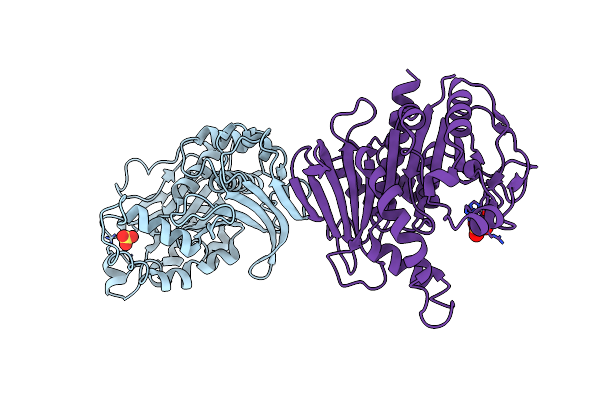
Crystal Structure Of Glutathionylated Glycolytic Glyceraldehyde-3- Phosphate Dehydrogenase From Arabidopsis Thaliana (Atgapc1)
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.99 Å Release Date: 2019-12-04 Classification: OXIDOREDUCTASE Ligands: NAD, SO4, GSH |
 |
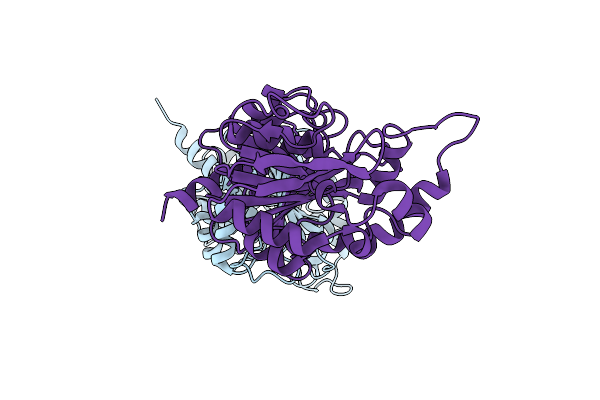
Crystal Structure Of Redox-Sensitive Phosphoribulokinase (Prk) From The Green Algae Chlamydomonas Reinhardtii
Organism: Chlamydomonas reinhardtii
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2019-04-10 Classification: PHOTOSYNTHESIS Ligands: SO4 |
 |
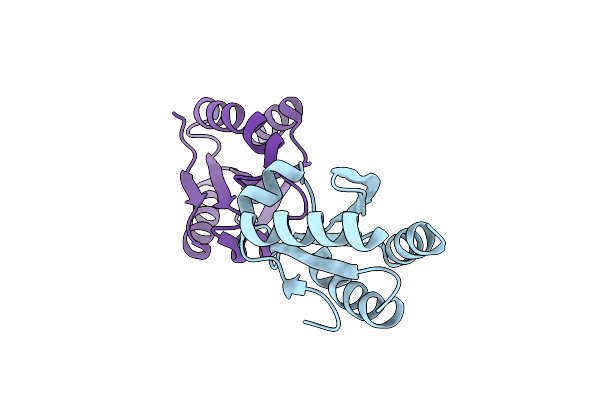
Crystal Structure Of Redox-Sensitive Phosphoribulokinase (Prk) From Arabidopsis Thaliana
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.47 Å Release Date: 2019-04-10 Classification: PHOTOSYNTHESIS |
 |
Organism: Chlamydomonas reinhardtii
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2019-01-16 Classification: ELECTRON TRANSPORT |
 |
Crystal Structure Of Partially Oxidized Thioredoxin H1 From Chlamydomonas Reinhardtii
Organism: Chlamydomonas reinhardtii
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2019-01-16 Classification: ELECTRON TRANSPORT |
 |
Crystal Structure (Orthorombic Form) Of C36S Mutant Of Thioredoxin H1 From Chlamydomonas Reinhardtii
Organism: Chlamydomonas reinhardtii
Method: X-RAY DIFFRACTION Resolution:0.94 Å Release Date: 2019-01-16 Classification: ELECTRON TRANSPORT Ligands: PEG |
 |
Crystal Structure Of C39S Mutant Of Thioredoxin H1 From Chlamydomonas Reinhardtii
Organism: Chlamydomonas reinhardtii
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2019-01-16 Classification: ELECTRON TRANSPORT |
 |
Crystal Structure (Trigonal Form) Of C36S Mutant Of Thioredoxin H1 From Chlamydomonas Reinhardtii
Organism: Chlamydomonas reinhardtii
Method: X-RAY DIFFRACTION Resolution:1.22 Å Release Date: 2019-01-16 Classification: ELECTRON TRANSPORT Ligands: PEG |
 |
Crystal Structure Of Chloroplastic Triosephosphate Isomerase From Chlamydomonas Reinhardtii At 1.1 A Of Resolution
Organism: Chlamydomonas reinhardtii
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2014-01-01 Classification: ISOMERASE Ligands: MRD, MPD |

