Search Count: 15
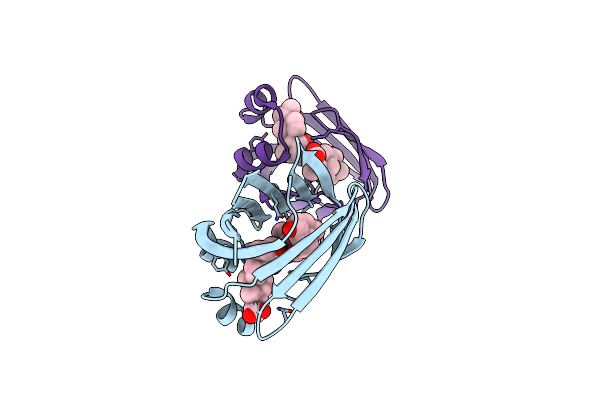 |
Crystal Structure Of Recombinant Chicken Liver Bile Acid Binding Protein (Cl-Babp) In Complex With Chenodeoxycholic Acid
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2024-10-09 Classification: LIPID BINDING PROTEIN Ligands: JN3 |
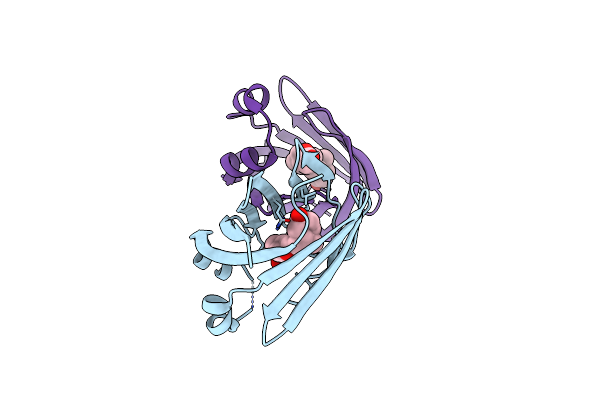 |
Crystal Structure Of Recombinant Chicken Liver Bile Acid Binding Protein (Cl-Babp) In Complex With Ursodeoxycholic Acid
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2024-10-09 Classification: LIPID BINDING PROTEIN Ligands: IU5 |
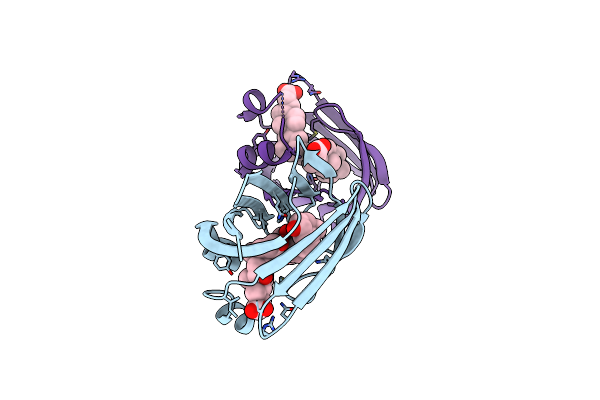 |
Crystal Structure Of Recombinant Chicken Liver Bile Acid Binding Protein (Cl-Babp) In Complex With Deoxycholic Acid
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-10-09 Classification: LIPID BINDING PROTEIN Ligands: DXC |
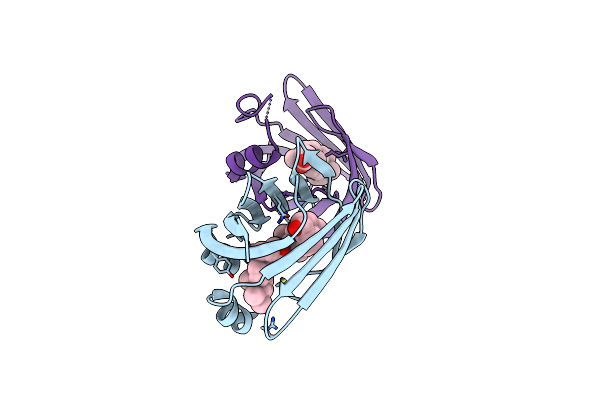 |
Crystal Structure Of Recombinant Chicken Liver Bile Acid Binding Protein (Cl-Babp) In Complex With Lithocholic Acid
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-10-09 Classification: LIPID BINDING PROTEIN Ligands: 4OA |
 |
Crystal Structure Of Recombinant Chicken Liver Bile Acid Binding Protein (Cl-Babp) In Complex With Ca-M11
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-10-09 Classification: LIPID BINDING PROTEIN Ligands: A1H7S |
 |
Complex Crystal Structure Of Mutant Human Monoglyceride Lipase With Compound 5D
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2024-01-31 Classification: HYDROLASE Ligands: F28, EDO |
 |
Complex Crystal Structure Of Mutant Human Monoglyceride Lipase With Compound 5L
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2024-01-31 Classification: HYDROLASE Ligands: EJI, EDO |
 |
Complex Crystal Structure Of Mutant Human Monoglyceride Lipase With Compound 5R
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2024-01-31 Classification: HYDROLASE Ligands: EFH, EDO |
 |
Structure Of Human Heat Shock Protein 90-Alpha N-Terminal Domain (Hsp90-Ntd) In Complex With Jmc31
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2022-09-07 Classification: CHAPERONE Ligands: M0U |
 |
Structure Of Human Heat Shock Protein 90-Alpha N-Terminal Domain (Hsp90-Ntd) Variant K112A In Complex With Jmc31
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.32 Å Release Date: 2022-09-07 Classification: CHAPERONE Ligands: M0U |
 |
Structure Of Human Heat Shock Protein 90-Alpha N-Terminal Domain (Hsp90-Ntd) Variant K112R In Complex With Jmc31
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2022-09-07 Classification: CHAPERONE Ligands: M0U |
 |
Crystal Structure Of Danio Rerio Histone Deacetylase 6 Catalytic Domain 2 (Cd2) Complexed With Nf2376
Organism: Danio rerio
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2020-12-02 Classification: HYDROLASE Ligands: ZN, K, QQD, EDO |
 |
Crystal Structure Of The Kainate Receptor Gluk3 Ligand Binding Domain In Complex With (S)-1-[2'-Amino-2'-Carboxyethyl]-6-Methyl-5,7-Dihydropyrrolo[3,4-D]Pyrimidin-2,4(1H,3H)-Dione At Resolution 2.4A
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2018-02-28 Classification: MEMBRANE PROTEIN Ligands: ZN, CG8, CL, SO4 |
 |
Crystal Structure Of The Kainate Receptor Gluk3 Ligand Binding Domain In Complex With (S)-1-[2-Amino-2-Carboxyethyl]-5,7-Dihydrothieno[3,4-D]Pyrimidin-2,4(1H,3H)-Dione At Resolution 2.6A
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2018-02-28 Classification: MEMBRANE PROTEIN Ligands: CGW, K, CL |
 |
Crystal Structure Of The Ligand Binding Domain Of Gluk1 In Complex With An Antagonist (S)-1-(2'-Amino-2'-Carboxyethyl)-3-[(2-Carboxythien-3-Yl)Methyl]Thieno[3,4-D]Pyrimidin-2,4-Dione At 2.5 A Resolution
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2011-06-22 Classification: MEMBRANE PROTEIN Ligands: 3HU, SO4, GOL, CL |

