Search Count: 30
 |
Organism: Fuscovulum blasticum dsm 2131
Method: ELECTRON MICROSCOPY Resolution:2.84 Å Release Date: 2025-03-05 Classification: PHOTOSYNTHESIS Ligands: BCL, SPO, PC1, CDL, BPH, U10, FE2 |
 |
Organism: Fuscovulum blasticum dsm 2131
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: PHOTOSYNTHESIS Ligands: BCL, SPO, PC1, CDL, BPH, U10, FE2 |
 |
Organism: Priestia megaterium
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2023-03-08 Classification: OXIDOREDUCTASE Ligands: HEM, EDO, SO4, BTB |
 |
Cryo-Em Structure Of C-Mannosyltransferase Cedpy19, In Complex With Acceptor Peptide And Bound To Cmt2-Fab And Anti-Fab Nanobody
Organism: Synthetic construct, Caenorhabditis elegans
Method: ELECTRON MICROSCOPY Release Date: 2023-01-11 Classification: MEMBRANE PROTEIN |
 |
Cryo-Em Structure Of C-Mannosyltransferase Cedpy19, In Apo State, Bound To Cmt2-Fab And Anti-Fab Nanobody
Organism: Synthetic construct, Caenorhabditis elegans
Method: ELECTRON MICROSCOPY Release Date: 2023-01-11 Classification: MEMBRANE PROTEIN |
 |
Cryo-Em Structure Of C-Mannosyltransferase Cedpy19, In Complex With Dol25-P-Man And Bound To Cmt2-Fab And Anti-Fab Nanobody
Organism: Synthetic construct, Caenorhabditis elegans
Method: ELECTRON MICROSCOPY Release Date: 2023-01-11 Classification: MEMBRANE PROTEIN Ligands: IZY |
 |
Cryo-Em Structure Of C-Mannosyltransferase Cedpy19, In Ternary Complex With Dol25-P-C-Man And Acceptor Peptide, Bound To Cmt2-Fab And Anti-Fab Nanobody
Organism: Synthetic construct, Caenorhabditis elegans
Method: ELECTRON MICROSCOPY Release Date: 2023-01-11 Classification: MEMBRANE PROTEIN Ligands: IZU |
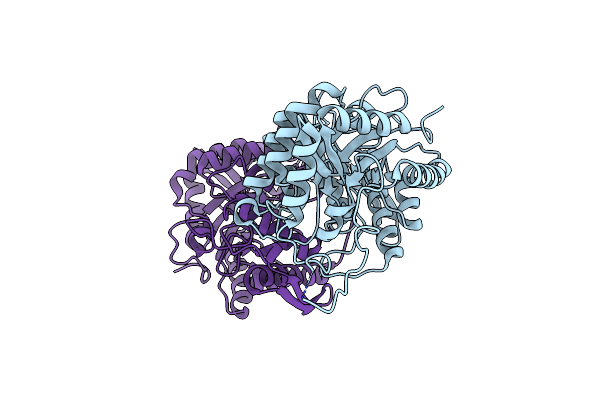 |
Crystal Structure Of Fmnh2-Dependent Monooxygenase From Agrobacterium Tumefaciens For Oxidative Desulfurization Of Sulfoquinovose
Organism: Agrobacterium fabrum (strain c58 / atcc 33970)
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2022-02-02 Classification: OXIDOREDUCTASE |
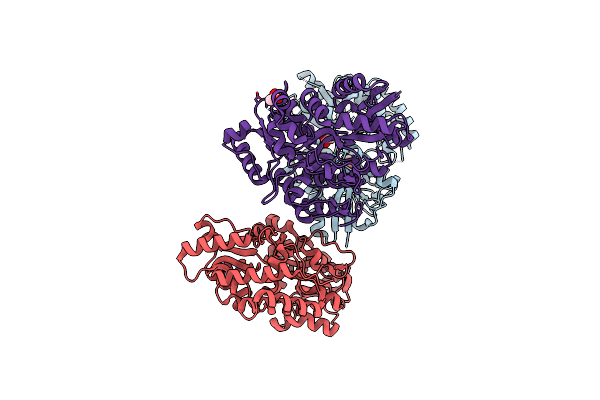 |
Crystal Structure Of Ligand Free Open Conformation Of Sulfoquinovosyl Binding Protein (Sqbp) From Agrobacterium Tumefaciens
Organism: Rhizobium radiobacter
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2022-01-19 Classification: SUGAR BINDING PROTEIN Ligands: ACT |
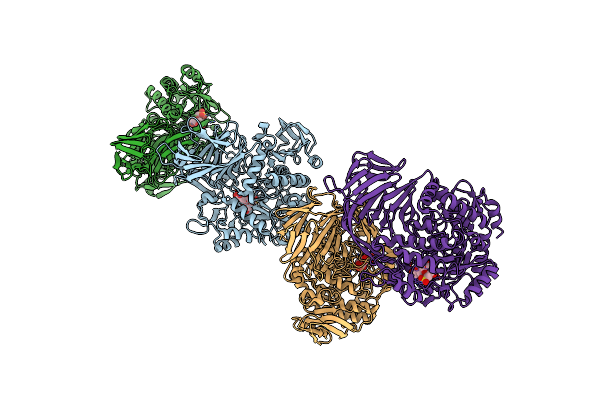 |
Crystal Structure Of A Gh31 Family Sulfoquinovosidase Mutant D455N From Agrobacterium Tumefaciens In Complex With Sulfoquinovosyl Glycerol (Sqgro)
Organism: Rhizobium radiobacter
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2022-01-19 Classification: HYDROLASE Ligands: VCW |
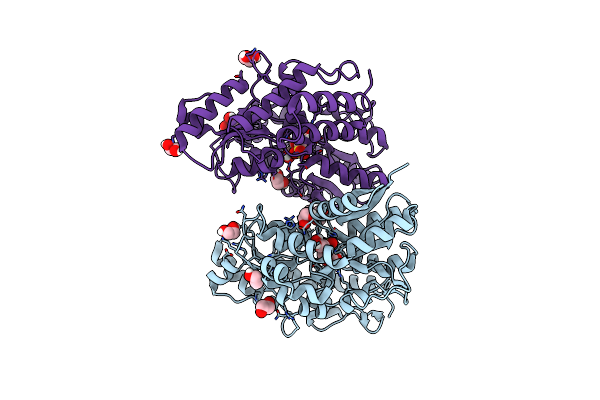 |
Crystal Structure Of Sq Binding Protein From Agrobacterium Tumefaciens In Complex With Sulfoquinovosyl Glycerol (Sqgro)
Organism: Rhizobium radiobacter
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2022-01-19 Classification: SUGAR BINDING PROTEIN Ligands: VCW, EDO |
 |
Crystal Structure Of Fmnh2-Dependent Monooxygenase For Oxidative Desulfurization Of Sulfoquinovose
Organism: Rhizobium oryzae
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2022-01-19 Classification: OXIDOREDUCTASE |
 |
Crystal Structure Of Aldo-Keto Reductase With C-Terminal His Tag From Agrobacterium Tumefaciens
Organism: Agrobacterium fabrum (strain c58 / atcc 33970)
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2022-01-12 Classification: OXIDOREDUCTASE Ligands: PO4 |
 |
Crystal Structure Of Apo Aldo-Keto Reductase From Agrobacterium Tumefaciens
Organism: Agrobacterium fabrum (strain c58 / atcc 33970)
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2022-01-12 Classification: OXIDOREDUCTASE |
 |
Crystal Structure Of Aldo-Keto Reductase From Agrobacterium Tumefaciens In A Binary Complex With Nadph
Organism: Agrobacterium fabrum (strain c58 / atcc 33970)
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2022-01-12 Classification: OXIDOREDUCTASE Ligands: PO4 |
 |
Crystal Structure Of Aldo-Keto Reductase From Agrobacterium Tumefaciens In A Ternary Complex With Nadph And Glucose
Organism: Agrobacterium fabrum (strain c58 / atcc 33970)
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2022-01-12 Classification: OXIDOREDUCTASE Ligands: NAP, GLC |
 |
Organism: Homo sapiens, Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2020-07-08 Classification: IMMUNE SYSTEM Ligands: PO4, MAN |
 |
Crystal Structure Of Human Alpha 1,6-Fucosyltransferase, Fut8 Bound To Gdp And A2Sgp
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2020-02-26 Classification: TRANSFERASE Ligands: ASN, GDP, SO4 |
 |
Crystal Structure Of Human Alpha 1,6-Fucosyltransferase, Fut8 In Its Apo-Form
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2020-02-26 Classification: TRANSFERASE Ligands: SO4 |
 |
Crystal Structure Of Mouse Alpha 1,6-Fucosyltransferase, Fut8 In Its Apo-Form
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-02-26 Classification: TRANSFERASE Ligands: SO4, EDO |

