Search Count: 18
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2018-09-19 Classification: DE NOVO PROTEIN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2018-09-19 Classification: DE NOVO PROTEIN Ligands: 38E |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2018-09-19 Classification: DE NOVO PROTEIN Ligands: 38E |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2018-09-19 Classification: DE NOVO PROTEIN Ligands: SO4 |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2018-09-19 Classification: DE NOVO PROTEIN |
 |
Sparse-Restraint Solution Nmr Structure Of Micelle-Solubilized Cytosolic Amino Terminal Domain Of C. Elegans Mechanosensory Ion Channel Mec-4 Refined By Restrained Rosetta
Organism: Caenorhabditis elegans
Method: SOLUTION NMR Release Date: 2017-02-01 Classification: TRANSPORT PROTEIN |
 |
Restrained Cs-Rosetta Solution Nmr Structure Of Staphylococcus Aureus Protein Sav1430. Northeast Structural Genomics Target Zr18. Structure Determination
Organism: Staphylococcus aureus subsp. aureus
Method: SOLUTION NMR Release Date: 2013-08-21 Classification: UNKNOWN FUNCTION |
 |
Restrained Cs-Rosetta Solution Nmr Structure Of The Cardb Domain Of Pf1109 From Pyrococcus Furiosus. Northeast Structural Genomics Consortium Target Pfr193A
Organism: Pyrococcus furiosus
Method: SOLUTION NMR Release Date: 2013-08-21 Classification: UNKNOWN FUNCTION |
 |
Refined Solution Nmr Structure Of Staphylococcus Aureus Protein Sav1430. Northeast Strucutral Genomics Consortium Target Zr18
Organism: Staphylococcus aureus subsp. aureus
Method: SOLUTION NMR Release Date: 2013-05-01 Classification: UNKNOWN FUNCTION |
 |
Solution Nmr Structure Of De Novo Designed Protein, P-Loop Ntpase Fold, Northeast Structural Genomics Consortium Target Or28
Organism: Artificial gene
Method: SOLUTION NMR Release Date: 2011-01-12 Classification: DE NOVO PROTEIN |
 |
Solution Nmr Structure Of Sap Domain Of Mkl/Myocardin-Like Protein 1 From H.Sapiens, Northeast Structural Genomics Consortium Target Hr4547E
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2010-06-16 Classification: TRANSCRIPTION REGULATOR |
 |
Solution Nmr Structure Of Sap Domain Of Mkl/Myocardin-Like Protein 1 From H.Sapiens, Northeast Structural Genomics Consortium Target Target Hr4547E
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2010-04-21 Classification: Transcription regulator |
 |
Solution Nmr Of Putative Excisionase From Klebsiella Pneumoniae, Northeast Structural Genomics Consortium Target Target Kpr49
Organism: Klebsiella pneumoniae
Method: SOLUTION NMR Release Date: 2010-04-14 Classification: HYDROLASE |
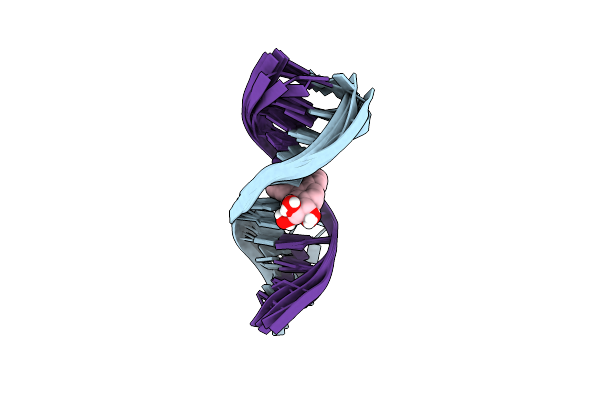 |
Solution Conformation Of The (+)Trans-Anti-Benzo[G]Chrysene-Da Adduct Opposite Dt In A Dna Duplex
|
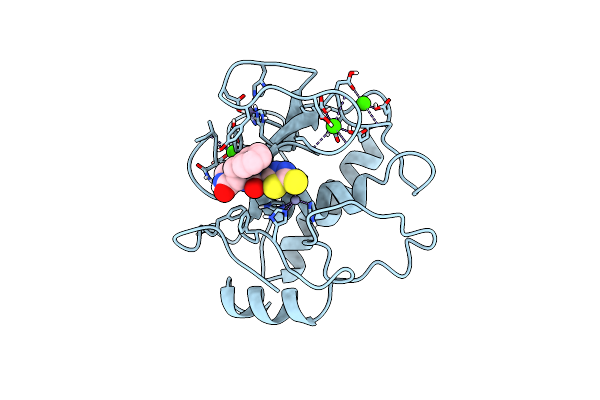 |
Structure Of The Catalytic Domain Of Human Fibroblast Stromelysin-1 Inhibited With The Thiadiazole Inhibitor Ipnu-107859, Nmr, 1 Structure
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 1999-01-20 Classification: HYDROLASE Ligands: ZN, CA, ATT |
 |
Solution Structure Of The [Af]-C8-Dg Adduct Opposite A-2 Deletion Site In The Nari Hot Spot Sequence Context; Nmr, 6 Structures
|
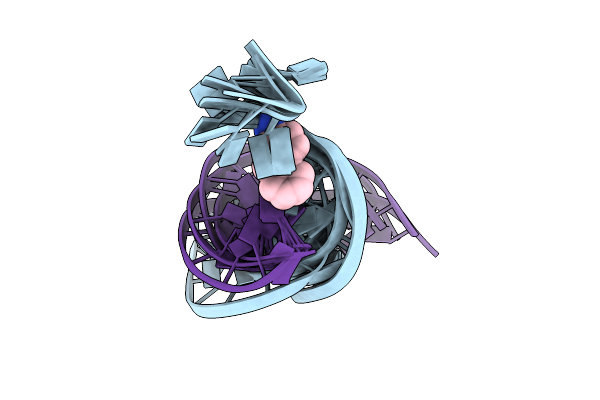 |
Solution Structure Of The [Af]-C8-Dg Adduct Positioned At A Template-Primer Junction, Nmr, 6 Structures
|
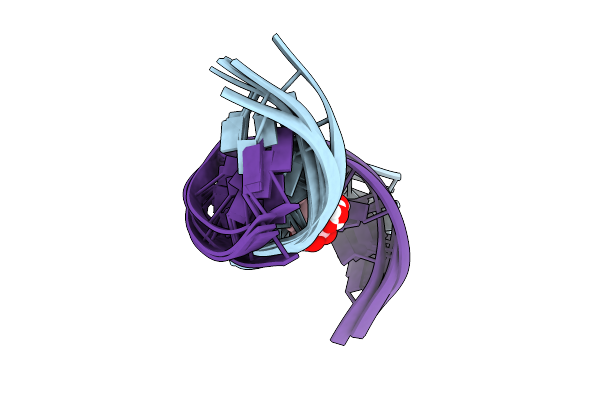 |
Solution Nmr Structure Of The [Bp]Da Adduct Opposite Dt In A Dna Duplex, 6 Structures
|

