Search Count: 24
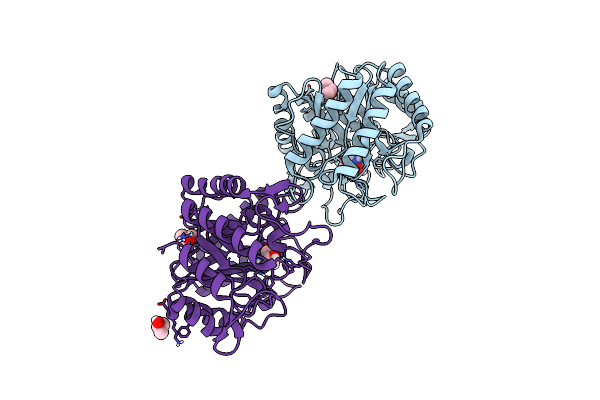 |
Structure Of Thermotoga Maritima Gh5 Endoglucanase Tm1752 In Complex With Tris
Organism: Thermotoga maritima (strain atcc 43589 / msb8 / dsm 3109 / jcm 10099)
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2022-03-30 Classification: HYDROLASE Ligands: TRS, IPA |
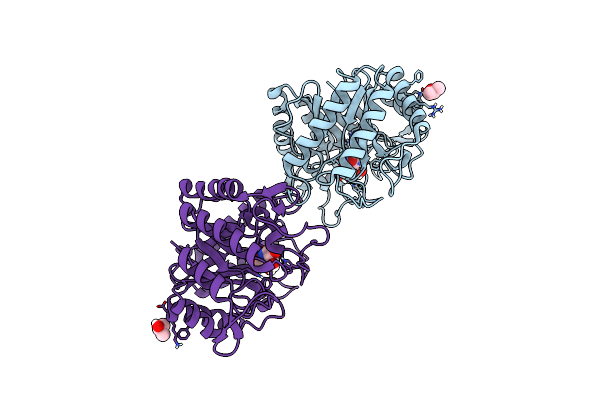 |
Structure Of The Thermotoga Maritima Family 5 Endo-Glucanase In Complex With 1-Deoxynojiromycin
Organism: Thermotoga maritima (strain atcc 43589 / msb8 / dsm 3109 / jcm 10099)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2022-03-16 Classification: HYDROLASE Ligands: NOJ, IPA |
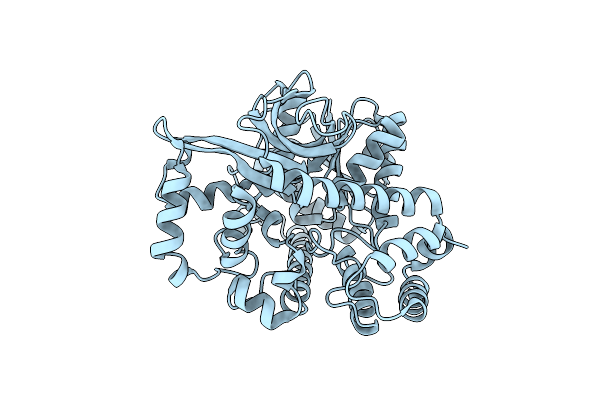 |
Crystal Structure Of Apo Form Of Alpha-Glucuronidase (Tm0752) From Thermotoga Maritima
Organism: Thermotoga maritima (strain atcc 43589 / msb8 / dsm 3109 / jcm 10099)
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2021-09-01 Classification: HYDROLASE |
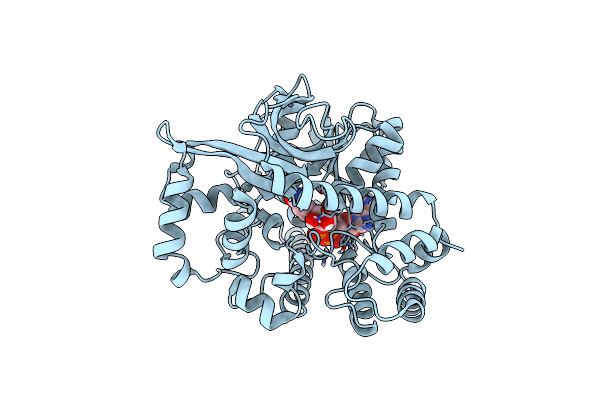 |
Crystal Structure Of Nadh Bound Holo Form Of Alpha-Glucuronidase (Tm0752) From Thermotoga Maritima At 1.97 Angstrom Resolution
Organism: Thermotoga maritima (strain atcc 43589 / msb8 / dsm 3109 / jcm 10099)
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2021-09-01 Classification: HYDROLASE Ligands: NAI |
 |
Crystal Structure Of Thermotoga Maritima Alpha-Glucuronidase (Tm0752) In Complex With Nadh And D-Glucuronic Acid
Organism: Thermotoga maritima (strain atcc 43589 / msb8 / dsm 3109 / jcm 10099)
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2021-09-01 Classification: HYDROLASE Ligands: NAI, BDP, IPA |
 |
Structure Of Deletion Mutant Of Alpha-Glucuronidase (Tm0752) From Thermotoga Maritima
Organism: Thermotoga maritima (strain atcc 43589 / msb8 / dsm 3109 / jcm 10099)
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2021-03-31 Classification: HYDROLASE Ligands: MN, NAI |
 |
Structure Of Nadh Complex Of Thermotoga Maritima Alpha-Glucuronidase At 2.15 Angstrom Resolution
Organism: Thermotoga maritima (strain atcc 43589 / msb8 / dsm 3109 / jcm 10099)
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2021-03-31 Classification: HYDROLASE Ligands: NAI |
 |
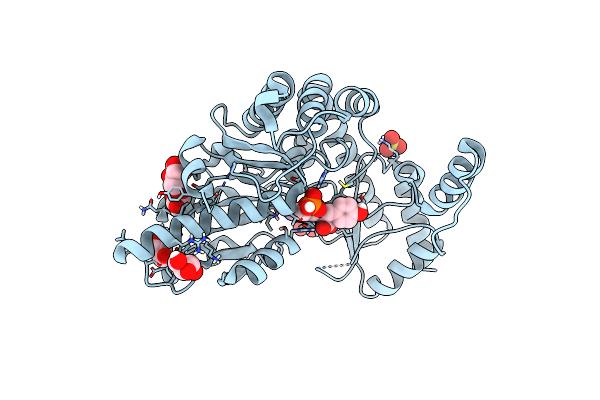
Structure Of A K245A Mutant Of A Group Ii Plp Dependent Decarboxylase From Methanocaldococcus Jannaschii, In Complex With Plp
Organism: Methanocaldococcus jannaschii (strain atcc 43067 / dsm 2661 / jal-1 / jcm 10045 / nbrc 100440)
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2020-12-02 Classification: LYASE Ligands: PLP, NH4, GOL, SO4 |
 |
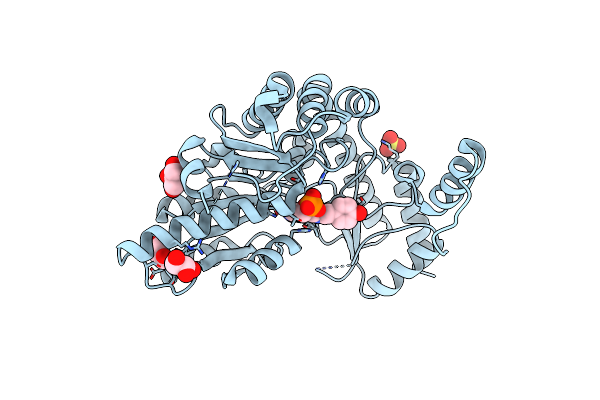
Structure Of A K245A Mutant Of L-Tyrosine Decarboxylase From Methanocaldococcus Jannaschii Complexed With L-Tyr: External Aldimine Form
Organism: Methanocaldococcus jannaschii (strain atcc 43067 / dsm 2661 / jal-1 / jcm 10045 / nbrc 100440)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-12-02 Classification: LYASE Ligands: SO4, GOL, 0PR |
 |
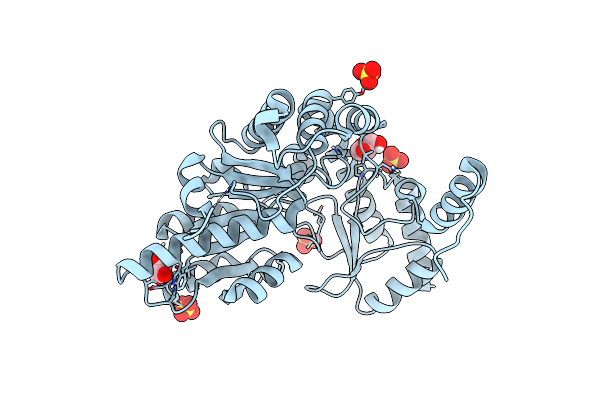
K245A Mutant Of L-Tyrosine Decarboxylase From Methanocaldococcus Jannaschii Complexed With A Post-Decarboxylation Quinonoid-Like Intermediate Formed With L-Tyrosine
Organism: Methanocaldococcus jannaschii (strain atcc 43067 / dsm 2661 / jal-1 / jcm 10045 / nbrc 100440)
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2020-12-02 Classification: LYASE Ligands: GOL, SO4, EBR |
 |
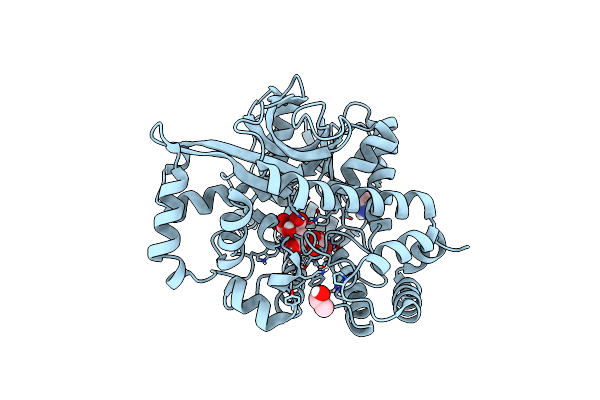
Structure Of A R371A Mutant Of A Group Ii Plp Dependent Decarboxylase From Methanocaldococcus Jannaschii
Organism: Methanocaldococcus jannaschii (strain atcc 43067 / dsm 2661 / jal-1 / jcm 10045 / nbrc 100440)
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2020-12-02 Classification: LYASE Ligands: SO4, GOL |
 |
Crystal Structure Of Citrate Complex Of Alpha-Glucuronidase (Tm0752)From Thermotoga Maritima
Organism: Thermotoga maritima (strain atcc 43589 / msb8 / dsm 3109 / jcm 10099)
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2020-04-29 Classification: HYDROLASE Ligands: CO, IMD, IPA, CIT |
 |
Crystal Structure Of A Group Ii Pyridoxal Dependent Decarboxylase, Llp-Bound Form From Methanocaldococcus Jannaschii At 1.72 A
Organism: Methanocaldococcus jannaschii dsm 2661
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2019-10-30 Classification: LYASE Ligands: SO4, GOL |
 |
Crystal Structure Of Apo Form Of Xylose Reductase From Debaryomyces Nepalensis
Organism: Debaryomyces nepalensis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2018-11-07 Classification: OXIDOREDUCTASE |
 |
Crystal Structure Of Xylose Reductase From Debaryomyces Nepalensis In Complex With Nadp-Dtt Adduct
Organism: Debaryomyces nepalensis
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2018-11-07 Classification: OXIDOREDUCTASE Ligands: NDP, DTT |
 |
Crystal Structure Of The P228A Variant Of Thermotoga Maritima Acetyl Esterase
Organism: Thermotoga maritima (strain atcc 43589 / msb8 / dsm 3109 / jcm 10099)
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2017-06-21 Classification: HYDROLASE Ligands: ACT |
 |
Crystal Structure Of The Thermotoga Maritima Acetyl Esterase (Tm0077) Complex With A Substrate Analog
Organism: Thermotoga maritima (strain atcc 43589 / msb8 / dsm 3109 / jcm 10099)
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2017-03-01 Classification: HYDROLASE Ligands: OIA |
 |
Crystal Structure Of A Loop Truncation Variant Of Thermotoga Maritima Acetyl Esterase Tm0077 (Apo Structure) At 2.75 Angstrom Resolution
Organism: Thermotoga maritima (strain atcc 43589 / msb8 / dsm 3109 / jcm 10099)
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2016-05-25 Classification: HYDROLASE |
 |
Crystal Structure Of The Monoclinic Form Of Thermotoga Maritima Acetyl Esterase Tm0077 (Apo Structure) At 1.76 Angstrom Resolution
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2016-04-27 Classification: HYDROLASE Ligands: CL, ACT |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.91 Å Release Date: 2004-03-23 Classification: LYASE Ligands: FMN |

