Search Count: 7
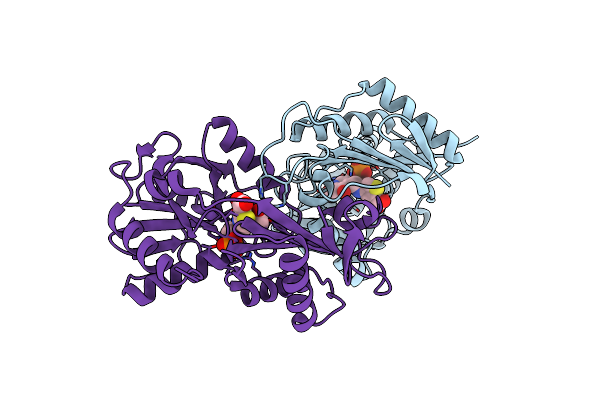 |
Complex Structure Of D-Amino Acid Aminotransferase And 4-Amino-4,5-Dihydro-Thiophenecarboxylic Acid (Adta)
Organism: Bacillus sp.
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2010-03-16 Classification: TRANSFERASE Ligands: PSZ, ACY |
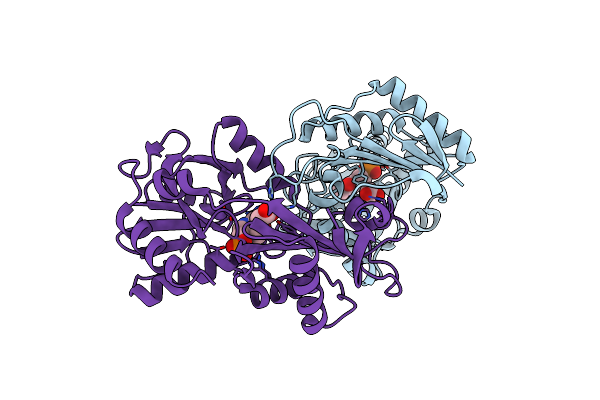 |
E177S Mutant Of The Pyridoxal-5'-Phosphate Enzyme D-Amino Acid Aminotransferase
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2000-11-15 Classification: TRANSFERASE Ligands: ACT, PLP |
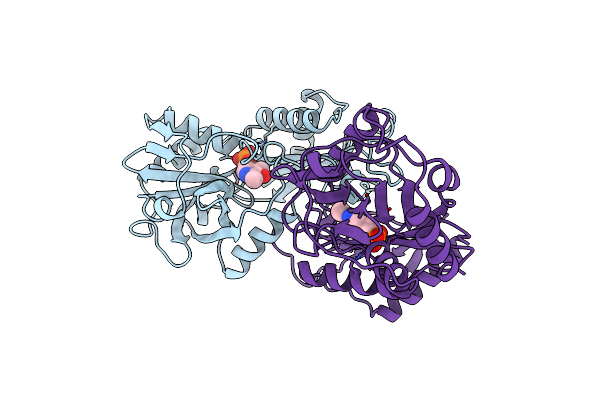 |
E177K Mutant Of D-Amino Acid Aminotransferase Complexed With Pyridoxamine-5'-Phosphate
Organism: Bacillus sp.
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 1998-12-14 Classification: TRANSFERASE Ligands: PLP |
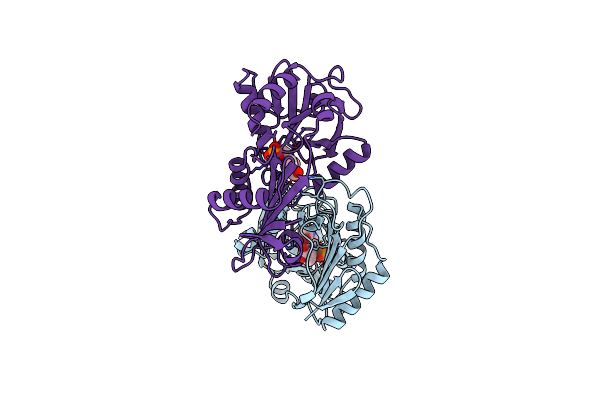 |
Crystallographic Structure Of D-Amino Acid Aminotransferase Inactivated By Pyridoxyl-D-Alanine
Organism: Bacillus sp.
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 1998-04-29 Classification: AMINOTRANSFERASE Ligands: PDD |
 |
Crystallographic Structure Of D-Amino Acid Aminotransferase In Pyridoxal-5'-Phosphate (Plp) Form
Organism: Bacillus sp.
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 1998-04-29 Classification: AMINOTRANSFERASE Ligands: SO4, PLP |
 |
Crystallographic Structure Of D-Amino Acid Aminotransferase Inactivated By D-Cycloserine
Organism: Bacillus sp.
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 1998-03-18 Classification: PYRIDOXAL PHOSPHATE Ligands: DCS |
 |
Crystallographic Structure Of D-Amino Acid Aminotransferase Complexed With Pyridoxal-5'-Phosphate
Organism: Bacillus sp.
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 1995-09-15 Classification: TRANSFERASE (AMINOTRANSFERASE) Ligands: PLP |

