Search Count: 364
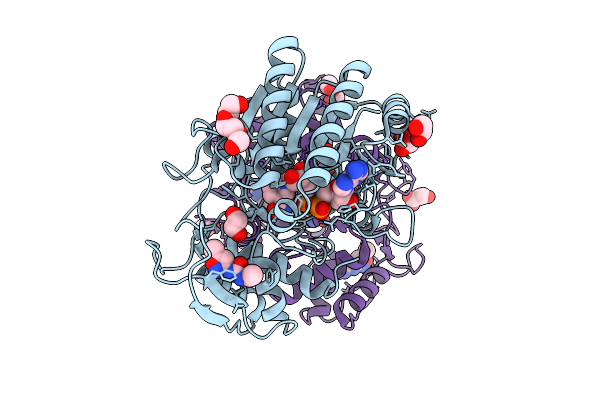 |
Structure Of Udp-Galactose-4-Epimerase (Gale) Bound To Fragment From Diamond Xchem Experiment.
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: CARBOHYDRATE Ligands: NAD, JGA, MLI, PGE, EDO, CL |
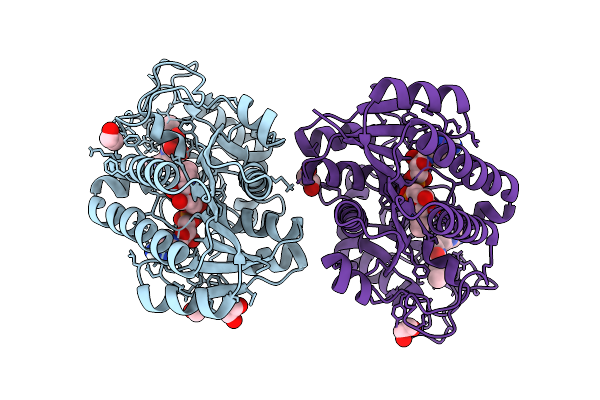 |
Structure Of Human Udp-Galactose 4-Epimerase In Complex With Compound Wbx04
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: ISOMERASE Ligands: EDO, NAD, A1IU2, MLT |
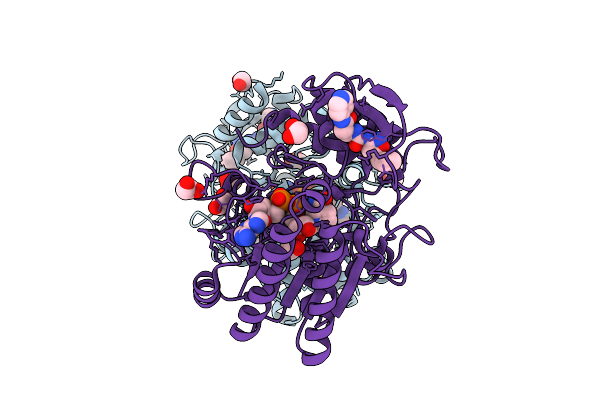 |
Structure Of Human Udp-Galactose 4-Epimerase In Complex With Compound Wbx09
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: ISOMERASE Ligands: EDO, NAD, A1IU5 |
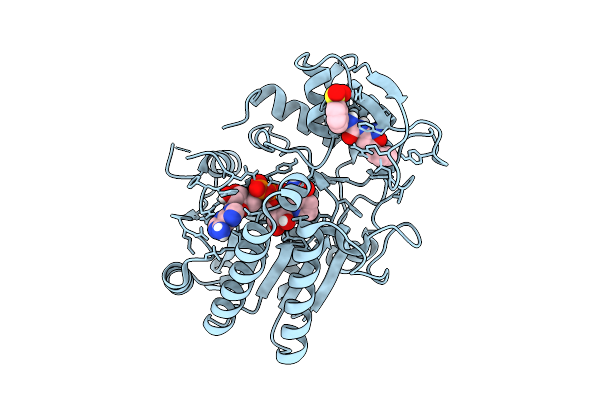 |
Structure Of Human Udp-Galactose 4-Epimerase In Complex With Covalent Compound Wbc10
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: ISOMERASE Ligands: NAI, A1IU6 |
 |
Organism: Synthetic construct, Arabidopsis thaliana
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: HYDROLASE/RNA Ligands: ZN, GOL |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: HYDROLASE Ligands: GOL, ZN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: LIGASE Ligands: A1CA9 |
 |
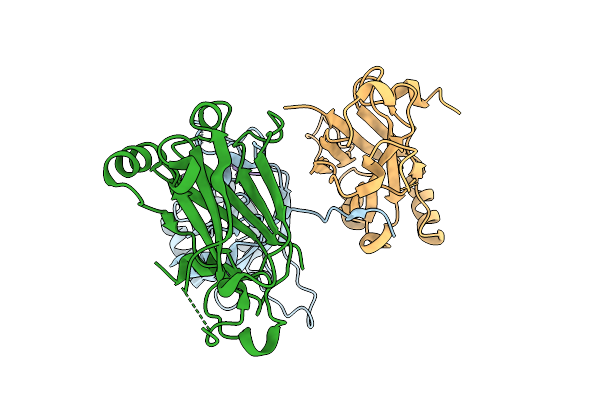
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: LIGASE Ligands: A1CA8 |
 |
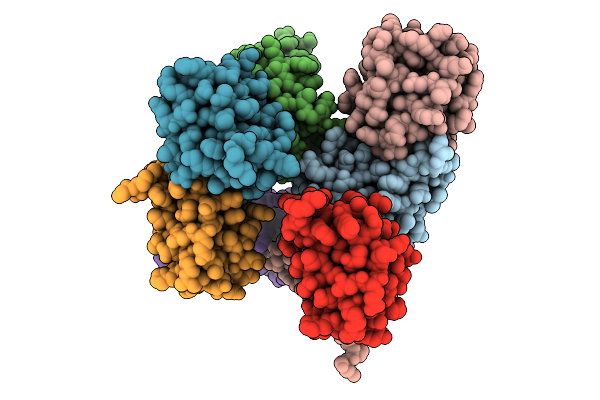 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: SIGNALING PROTEIN |
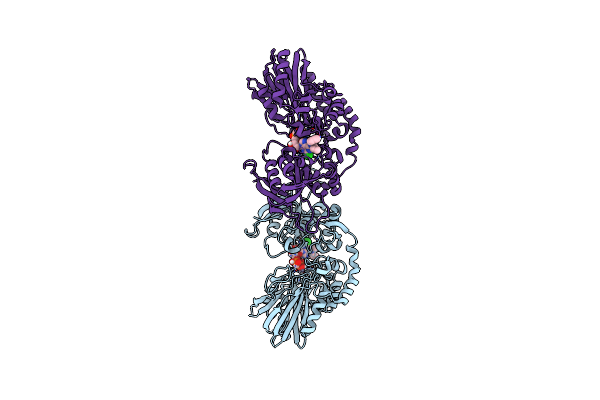 |
Crystal Structure Of Human Cd73 (Ecto-5'-Nucleotidase) In Complex With The Ampcp Analog Psb-12730 In The Closed State (Crystal Form Iv)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: HYDROLASE Ligands: ZN, A1IZZ |
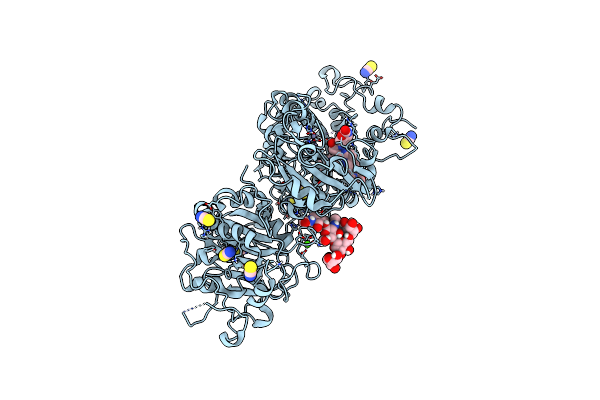 |
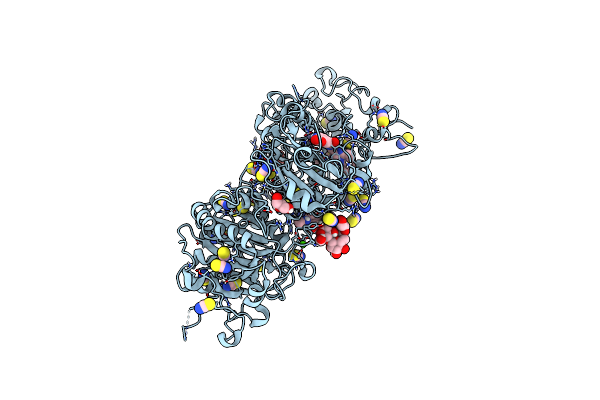
Crystal Structure Of Autotaxin (Enpp2) With Type Vi Inhibitor, A Novel Class Of Inhibitors With Three-Point Lock Binding Mode
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2025-04-23 Classification: HYDROLASE Ligands: ZN, CA, IOD, A1IG5, SCN |
 |
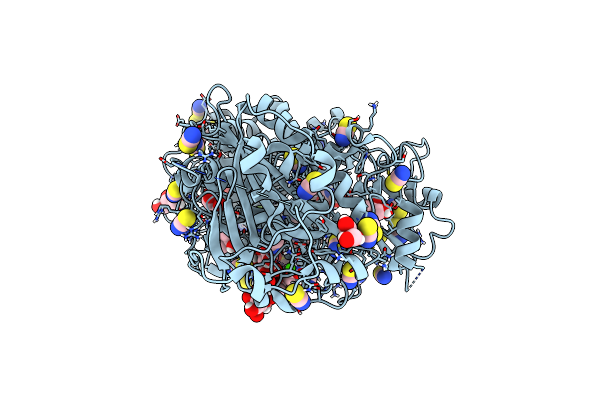
Crystal Structure Of Autotaxin (Enpp2) With Type Vi Inhibitor, A Novel Class Of Inhibitors With Three-Point Lock Binding Mode
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2025-04-23 Classification: HYDROLASE Ligands: A1IGZ, ZN, CA, IOD, SCN, GOL |
 |
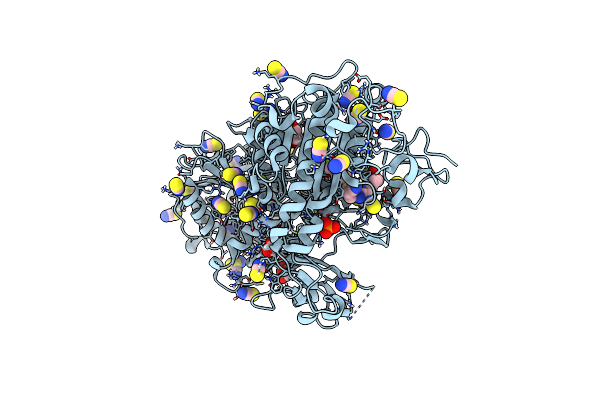
Crystal Structure Of Autotaxin (Enpp2) With Type Vi Inhibitor, A Novel Class Of Inhibitors With Three-Point Lock Binding Mode
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2025-04-23 Classification: HYDROLASE Ligands: ZN, CA, IOD, A1IG0, SCN, GOL |
 |
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-04-23 Classification: HYDROLASE Ligands: SCN, ZN, CA, IOD, A1IG1, GOL, PO4 |
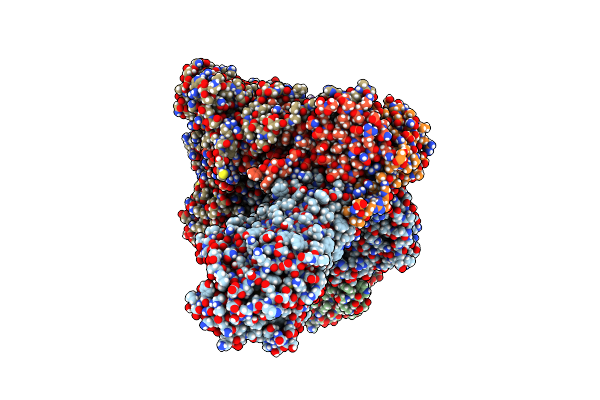 |
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-04-16 Classification: MEMBRANE PROTEIN |
 |
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-04-16 Classification: MEMBRANE PROTEIN |
 |
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-04-16 Classification: MEMBRANE PROTEIN |
 |
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-04-16 Classification: MEMBRANE PROTEIN |
 |
Organism: Escherichia coli k-12, Photorhabdus
Method: ELECTRON MICROSCOPY Release Date: 2025-04-16 Classification: MEMBRANE PROTEIN Ligands: MG |

