Search Count: 114
 |
Structure Of Active State Mc4R In Complex With A Potent Ligand Mimicking Nanobody
Organism: Homo sapiens, Lama glama, Escherichia coli, Discosoma sp.
Method: ELECTRON MICROSCOPY Release Date: 2024-09-25 Classification: MEMBRANE PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2024-09-04 Classification: TRANSFERASE Ligands: A1ADE, SO4, GDP |
 |
Crystal Structure Of The Tegument Protein Ul82 (Pp71) From Human Cytomegalovirus
Organism: Human betaherpesvirus 5
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2024-02-28 Classification: VIRAL PROTEIN Ligands: PG4 |
 |
Crystal Structure Of Histone H3 Lysine 79 (H3K79) Methyltransferase Rv2067C From Mycobacterium Tuberculosis
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2023-10-18 Classification: TRANSFERASE Ligands: PO4, SAH |
 |
T. Cruzi Topoisomerase Ii Alpha Bound To Dsdna And The Covalent Inhibitor Ct1
Organism: Trypanosoma cruzi strain cl brener, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2023-07-12 Classification: ISOMERASE Ligands: YWX |
 |
Crystal Structure Of Ap2 Associated Kinase 1 Complexed With 5-[(4-Aminopiperidin-1-Yl)Methyl]-N-{3-[5-(Propan-2-Yl)-1,3,4-Thiadiazol-2-Yl]Phenyl}Pyrrolo[2,1-F][1,2,4]Triazin-4-Amine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2023-07-12 Classification: TRANSFERASE Ligands: YFV, SO4 |
 |
Crystal Structure Of Ap2 Associated Kinase 1 Complexed With (5P)-3-({(8R)-5-[(4-Aminopiperidin-1-Yl)Methyl]Pyrrolo[2,1-F][1,2,4]Triazin-4-Yl}Amino)-5-[2-(Propan-2-Yl)-2H-Tetrazol-5-Yl]Phenol
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-07-12 Classification: TRANSFERASE Ligands: ZRR, SO4 |
 |
Organism: Mycobacterium tuberculosis h37rv
Method: ELECTRON MICROSCOPY Release Date: 2022-05-25 Classification: LYASE Ligands: PLP |
 |
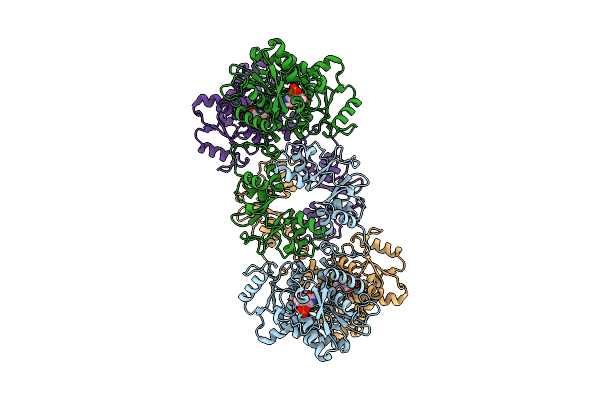
Cystathionine Beta-Synthase Of Mycobacterium Tuberculosis In The Presence Of S-Adenosylmethionine.
Organism: Mycobacterium tuberculosis h37rv
Method: ELECTRON MICROSCOPY Release Date: 2022-05-25 Classification: LYASE |
 |
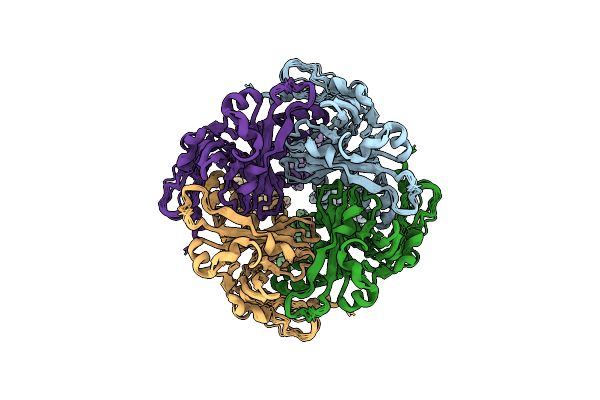
Cystathionine Beta-Synthase Of Mycobacterium Tuberculosis In The Presence Of S-Adenosylmethionine And Serine.
Organism: Mycobacterium tuberculosis h37rv
Method: ELECTRON MICROSCOPY Release Date: 2022-05-25 Classification: LYASE Ligands: PLS |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2022-05-18 Classification: STRUCTURAL PROTEIN Ligands: SO4, GOL, CL |
 |
3D Model Of The 3-Rbd Up Single Trimeric Spike Protein Of Sars-Cov2 In The Presence Of Synthetic Peptide Sih-5.
Organism: Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-04-27 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Crystal Structure Of Ap2 Associated Kinase 1 Isoform 1 Complexed With Ligand (2R)-2-Amino-N-[3-(Difluorom Ethoxy)-4-(1,3-Oxazol-5-Yl)Phenyl]-4-Methylpentanamide
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.13 Å Release Date: 2022-02-23 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: YFV, SO4, 5OI |
 |
Crystal Structure Of Ap2 Associated Kinase 1 Isoform 1 Complexed With Ligand 2-(1-Amino-3-Methylbutyl)-6- (Pyridin-4-Yl)Quinoline-4-Carbonitrile
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.12 Å Release Date: 2022-02-23 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: 5P6, SO4 |
 |
Crystal Structure Of Ap2 Associated Kinase 1 Isoform 1 Complexed With Ligand (2R)-2-Amino-N-[3-(Difluorom Ethoxy)-4-(1,3-Oxazol-5-Yl)Phenyl]-4-Methylpentanamide
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2022-02-23 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: 5QI, PG6, SO4, YFV |
 |
Organism: Burkholderia pseudomallei
Method: SOLID-STATE NMR Release Date: 2022-02-02 Classification: MEMBRANE PROTEIN |
 |
The Crystal Structure Of Q108K:K40E:T53A:R58W:Q38F:Q4F:Y19W Mutant Of Hcrbpii Bound With Lizfluor Chromophore Showing Excited State Intermolecular Proton Transfer
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2022-01-12 Classification: LIPID BINDING PROTEIN Ligands: ACT, ZFG |
 |
The Crystal Structure Of Q108K:K40H:T53A:R58L:Q38F:Q4F Mutant Of Hcrbpii Bound With Fr1 Chromophore Showing Excited State Intermolecular Proton Transfer
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.59 Å Release Date: 2022-01-12 Classification: LIPID BINDING PROTEIN Ligands: ZFJ, GOL |
 |
The Crystal Structure Of Q108K:K40L:T51V:T53S:R58W:Y19W:A33W:L117E Mutant Of Hcrbpii Bound With Lizfluor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.26 Å Release Date: 2022-01-12 Classification: LIPID BINDING PROTEIN Ligands: ACT, ZFG, GOL |
 |
The Crystal Structure Of Domain-Swapped Trimer Q108K:K40D:T53A:R58L:Q38F:Q4F Mutant Of Hcrbpii Bound With Lizfluor3 Chromophore Showing Excited State Intermolecular Proton Transfer
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2022-01-12 Classification: LIPID BINDING PROTEIN Ligands: ZFP, ACT, GOL |

