Search Count: 24
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2024-11-06 Classification: HYDROLASE Ligands: A1IL0 |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2024-11-06 Classification: HYDROLASE Ligands: A1IL7, SO4, GOL, EDO |
 |
Organism: Hepatitis b virus
Method: X-RAY DIFFRACTION Resolution:2.63 Å Release Date: 2024-09-25 Classification: VIRAL PROTEIN Ligands: A1AVK |
 |
Organism: Hepatitis b virus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2023-09-06 Classification: VIRAL PROTEIN Ligands: YWE, EDO |
 |
Cryo-Em Structure Of Ribosome-Sec61-Trap (Translocon Associated Protein) Translocon Complex
Organism: Oryctolagus cuniculus
Method: ELECTRON MICROSCOPY Release Date: 2023-03-01 Classification: MEMBRANE PROTEIN Ligands: MG, ZN |
 |
Cryo-Em Structure Of Ribosome-Sec61 In Complex With Cyclotriazadisulfonamide Derivative Ck147
Organism: Oryctolagus cuniculus
Method: ELECTRON MICROSCOPY Release Date: 2023-02-22 Classification: MEMBRANE PROTEIN Ligands: MG, ZN, OWI |
 |
Organism: Sinorhizobium meliloti 1021, Sinorhizobium meliloti
Method: X-RAY DIFFRACTION Resolution:2.72 Å Release Date: 2022-11-02 Classification: SIGNALING PROTEIN Ligands: CMP, MG |
 |
Organism: Sinorhizobium meliloti 1021, Sinorhizobium meliloti
Method: X-RAY DIFFRACTION Release Date: 2022-11-02 Classification: SIGNALING PROTEIN Ligands: PCG |
 |
Organism: Homo sapiens, Sus scrofa
Method: ELECTRON MICROSCOPY Resolution:3.89 Å Release Date: 2022-06-15 Classification: MOTOR PROTEIN Ligands: ANP, GTP, G2P, MG |
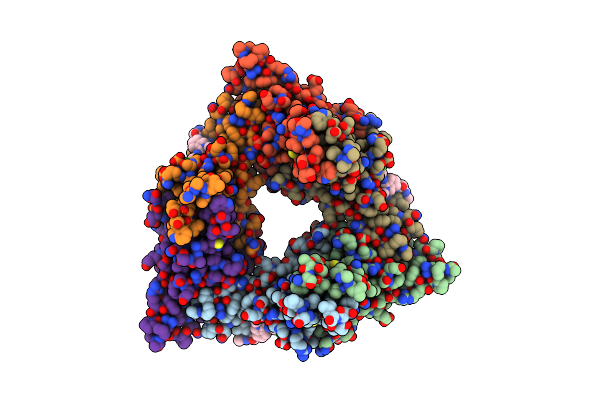 |
Organism: Hepatitis b virus genotype a
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2022-05-04 Classification: VIRAL PROTEIN/INHIBITOR Ligands: 8EI |
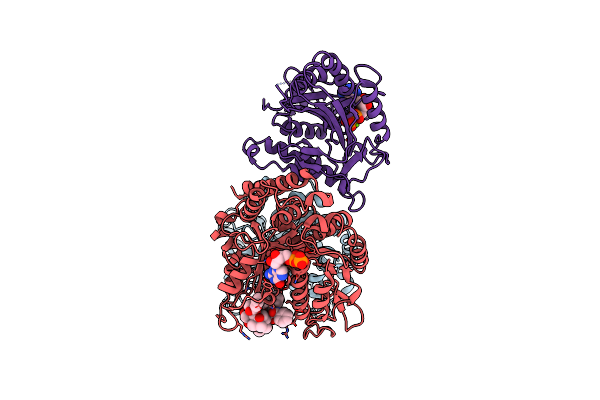 |
Organism: Homo sapiens, Sus scrofa
Method: ELECTRON MICROSCOPY Resolution:4.20 Å Release Date: 2019-05-01 Classification: MOTOR PROTEIN Ligands: MG, GTP, GDP, TA1, ADP |
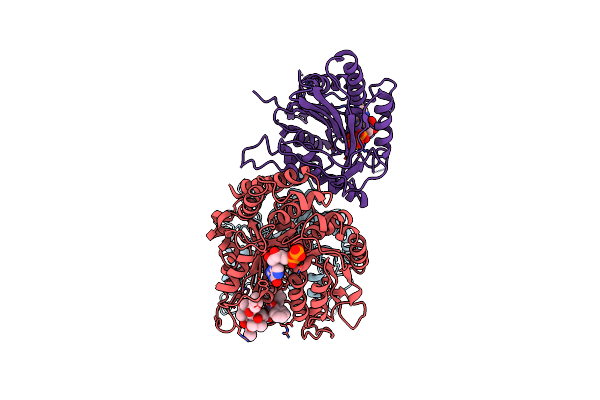 |
Organism: Homo sapiens, Sus scrofa
Method: ELECTRON MICROSCOPY Resolution:4.20 Å Release Date: 2019-05-01 Classification: motor protein/inhibitor Ligands: GTP, GDP, TA1, ANP |
 |
Organism: Ipomoea batatas
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2017-12-06 Classification: HYDROLASE Ligands: IPA |
 |
Crystal Structure Of Sweet Potato Beta-Amylase Complexed With Maltotetraose
Organism: Ipomoea batatas
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2017-12-06 Classification: HYDROLASE |
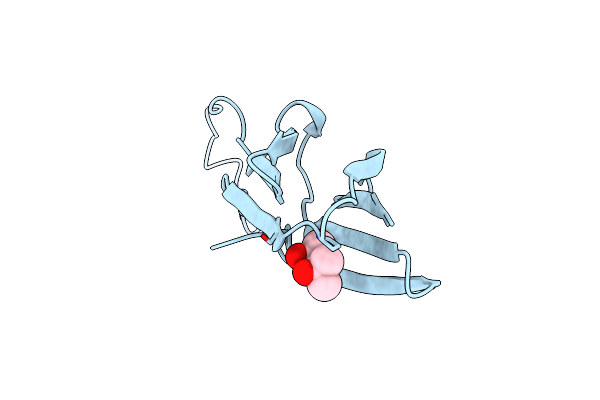 |
Small Heat Shock Protein Agsa From Salmonella Typhimurium: Alpha Crystallin Domain
Organism: Salmonella enterica subsp. enterica serovar typhimurium
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2016-04-20 Classification: CHAPERONE Ligands: MPD |
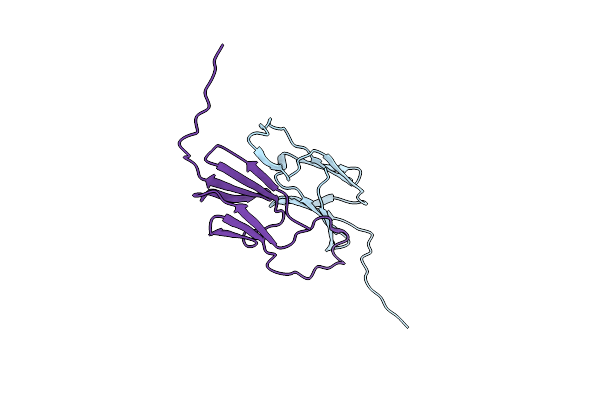 |
Small Heat Shock Protein Agsa From Salmonella Typhimurium: C-Terminal Truncated Construct
Organism: Salmonella enterica subsp. enterica serovar typhimurium
Method: X-RAY DIFFRACTION Resolution:4.10 Å Release Date: 2016-04-20 Classification: CHAPERONE |
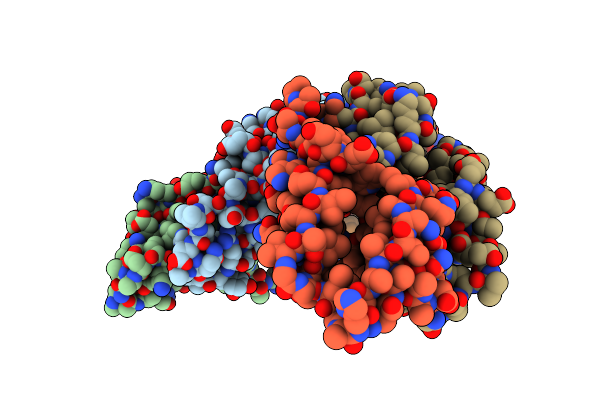 |
Small Heat Shock Protein Agsa From Salmonella Typhimurium: Truncations At N- And C- Termini
Organism: Salmonella enterica subsp. enterica serovar typhimurium
Method: X-RAY DIFFRACTION Resolution:7.50 Å Release Date: 2016-04-20 Classification: CHAPERONE |
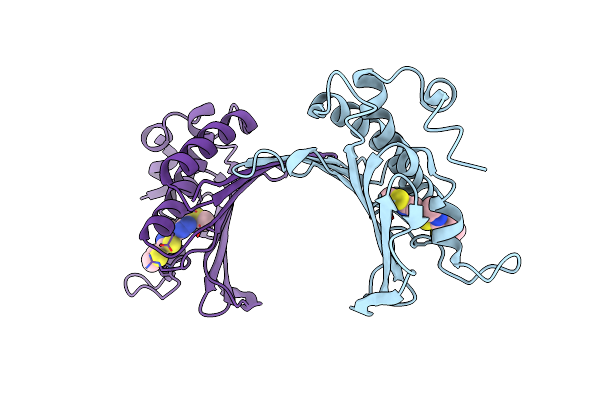 |
Crystal Structure Of Staphylococcus Aureus Gyrase B Co-Complexed With 4-Methyl-5-[3-(Methylsulfanyl)-1H-Pyrazol-5-Yl]-2-Thiophen-2-Yl-1,3-Thiazole Inhibitor
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2010-02-09 Classification: Isomerase/Isomerase inhibitor Ligands: B48 |
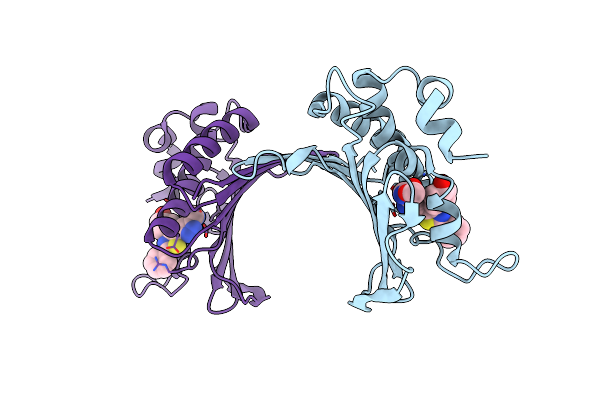 |
Staphylococcus Aureus Gyrase B Co-Complex With Methyl ({5-[4-(4-Hydroxypiperidin-1-Yl)-2-Phenyl-1,3-Thiazol-5-Yl]-1H-Pyrazol-3-Yl}Methyl)Carbamate Inhibitor
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2010-02-09 Classification: Isomerase/Isomerase inhibitor Ligands: B47 |
 |
Crystal Structure Of E. Coli Gyrase B Co-Complexed With Prop-2-Yn-1-Yl {[5-(4-Piperidin-1-Yl-2-Pyridin-3-Yl-1,3-Thiazol-5-Yl)-1H-Pyrazol-3-Yl]Methyl}Carbamate Inhibitor
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2010-02-09 Classification: Isomerase/Isomerase inhibitor Ligands: B46 |

