Search Count: 56
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: HYDROLASE Ligands: A1IW3, EDO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: HYDROLASE Ligands: A1IWL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: HYDROLASE Ligands: A1IWN |
 |
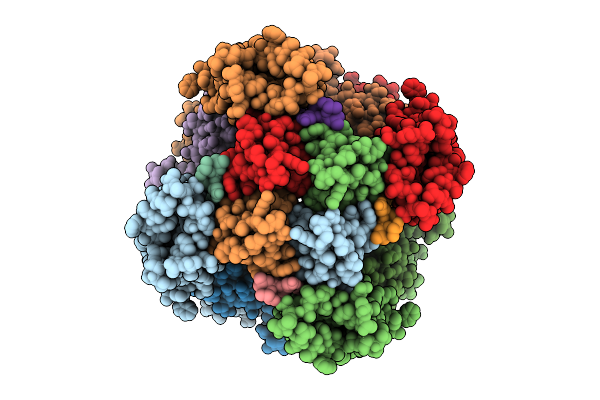
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: MEMBRANE PROTEIN Ligands: CA, A1BBG |
 |
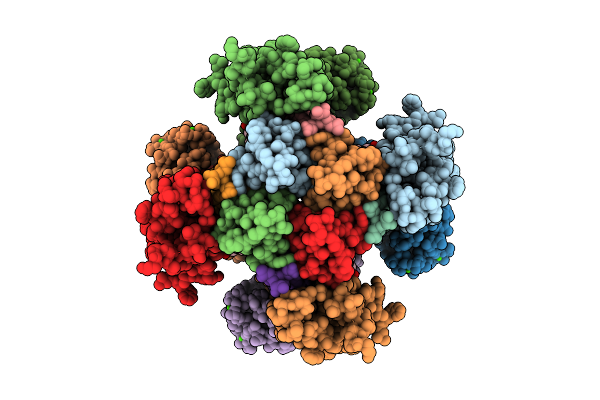
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-08-20 Classification: MEMBRANE PROTEIN Ligands: CA |
 |
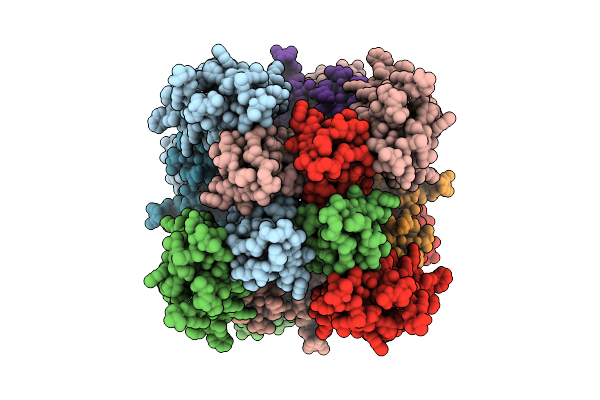
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-08-20 Classification: MEMBRANE PROTEIN Ligands: PT5, CA |
 |
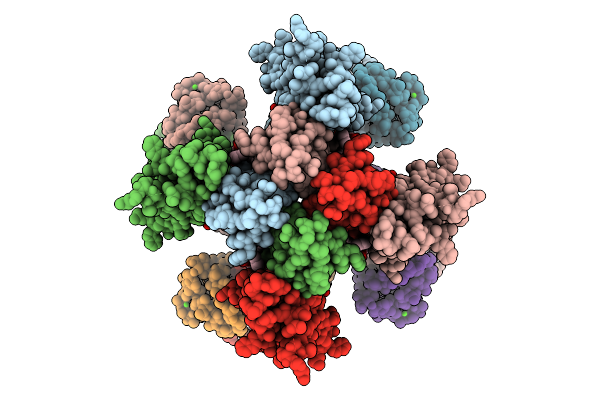
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-08-20 Classification: MEMBRANE PROTEIN Ligands: CA, PT5 |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-08-20 Classification: MEMBRANE PROTEIN Ligands: PT5, CA |
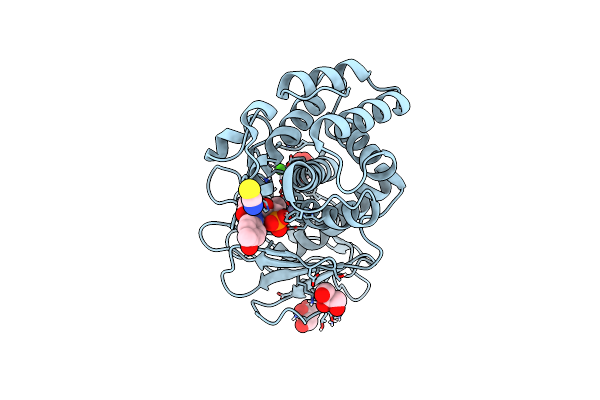 |
Crystal Structure Of Recombinant Lasb From Pseudomonas Aeruginosa Pao1 In Complex With 6466
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:1.31 Å Release Date: 2024-10-16 Classification: HYDROLASE Ligands: ZN, CA, GOL, SCN, XI5 |
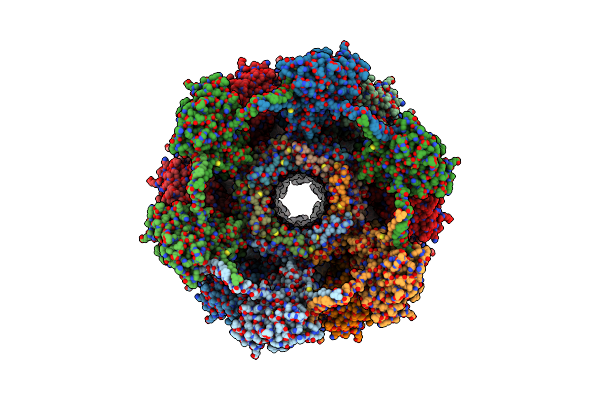 |
Organism: Clostridioides difficile
Method: ELECTRON MICROSCOPY Release Date: 2024-08-28 Classification: VIRUS LIKE PARTICLE |
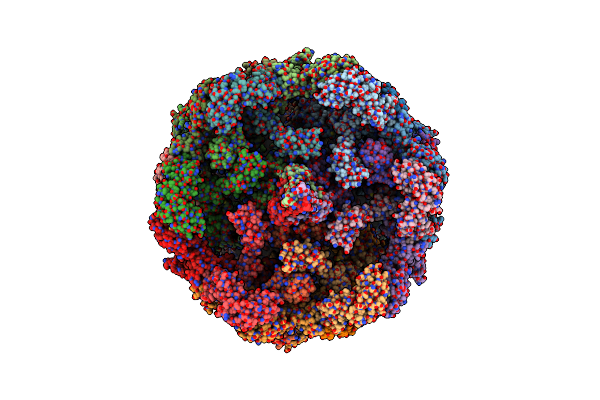 |
Cryoem Structure Of Diffocin - Precontracted - Baseplate - Focused Refinement On Triplex Region
Organism: Clostridioides difficile
Method: ELECTRON MICROSCOPY Release Date: 2024-08-28 Classification: VIRUS LIKE PARTICLE |
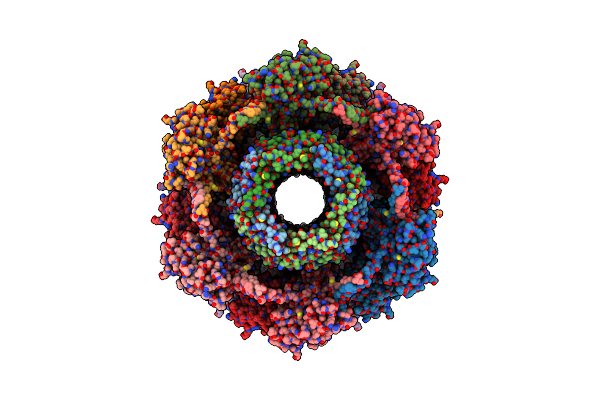 |
Organism: Clostridioides difficile
Method: ELECTRON MICROSCOPY Release Date: 2024-08-28 Classification: VIRUS LIKE PARTICLE |
 |
Organism: Clostridioides difficile
Method: ELECTRON MICROSCOPY Release Date: 2024-08-28 Classification: VIRUS LIKE PARTICLE |
 |
Cryoem Structure Of Diffocin - Postcontracted - Collar - Transitional State
Organism: Clostridioides difficile
Method: ELECTRON MICROSCOPY Release Date: 2024-08-28 Classification: VIRUS LIKE PARTICLE |
 |
Organism: Clostridioides difficile
Method: ELECTRON MICROSCOPY Release Date: 2024-08-28 Classification: VIRUS LIKE PARTICLE |
 |
Cryoem Structure Of Diffocin - Postcontracted - Baseplate - Transitional State
Organism: Clostridioides difficile
Method: ELECTRON MICROSCOPY Release Date: 2024-08-28 Classification: VIRUS LIKE PARTICLE |
 |
Organism: Clostridioides difficile
Method: ELECTRON MICROSCOPY Release Date: 2024-08-28 Classification: VIRUS LIKE PARTICLE |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-05-01 Classification: MEMBRANE PROTEIN Ligands: HEB, NDP, FAD, PX2, ZN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-05-01 Classification: MEMBRANE PROTEIN Ligands: HEB, NDP, FAD, D12, D10, ZN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-05-01 Classification: MEMBRANE PROTEIN Ligands: HEB, NDP, FAD, ZN, D12 |

