Search Count: 1,937
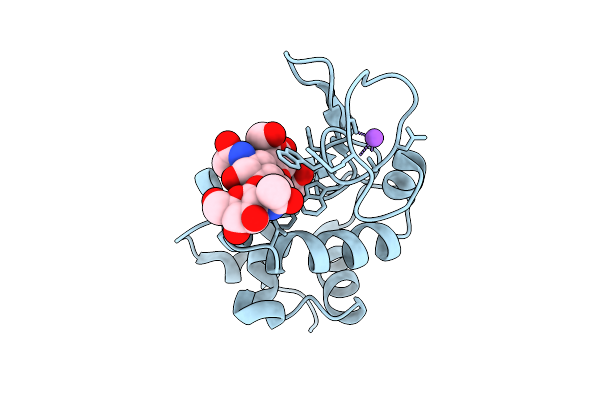 |
Structure Of Lysozyme-N,N',N"-Triacetylchitotriose Complex Determined Using Reyes For Automated Microed
Organism: Gallus gallus
Method: ELECTRON CRYSTALLOGRAPHY Release Date: 2025-12-10 Classification: HYDROLASE Ligands: CL, NA |
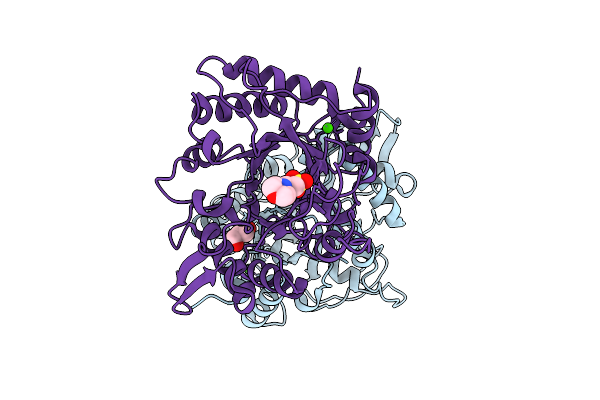 |
Organism: Listeria monocytogenes
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: HYDROLASE Ligands: GOL, CA, NA, MES |
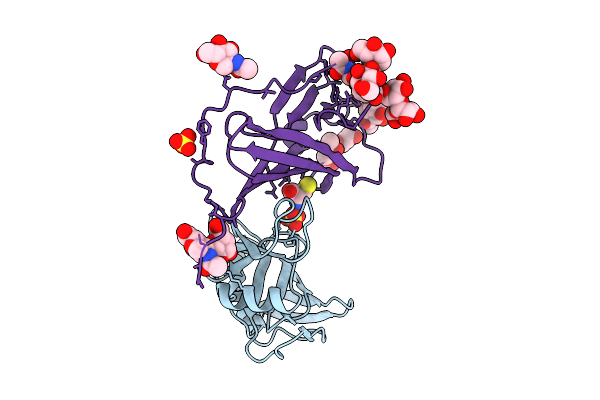 |
Organism: Mus musculus, Murid betaherpesvirus 1
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: VIRAL PROTEIN Ligands: SO4, NAG, CYS |
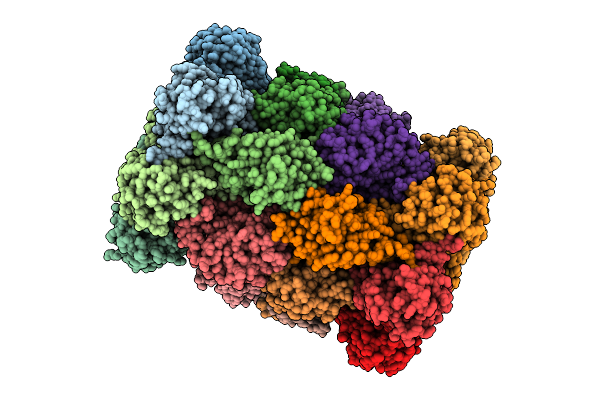 |
Organism: Plasmodium falciparum 3d7
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: CYTOSOLIC PROTEIN Ligands: A1B74 |
 |
Organism: Plasmodium falciparum 3d7
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: CYTOSOLIC PROTEIN Ligands: A1B73 |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: TRANSFERASE Ligands: A1CG3 |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: TRANSFERASE Ligands: A1CG7 |
 |
Organism: Rotavirus a
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: VIRAL PROTEIN |
 |
Organism: Rotavirus a
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: VIRAL PROTEIN |
 |
Structure Of Pou2F3 Pou Domains Bound To Coactivator Oca-T2 And Dna (2.8 Angstrom Resolution)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: TRANSCRIPTION |
 |
Structure Of Pou2F3 Pou Domains Bound To Coactivator Oca-T2 And Dna (2.1 Angstrom Resolution)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: TRANSCRIPTION |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: TRANSCRIPTION Ligands: BO3 |
 |
Organism: Xenopus laevis, Synthetic construct, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: NUCLEAR PROTEIN |
 |
Organism: Sars-cov-2 pseudovirus
Method: X-RAY DIFFRACTION Release Date: 2025-09-17 Classification: VIRAL PROTEIN Ligands: ZN, SAH, A1JLH, CL, IMD |
 |
Crystal Structure Of Ddb1-Crbn-Alv1 Complex Bound To Triple Znf Of Helios (Ikzf2 Zf1-3)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: LIGASE Ligands: RN9, ZN |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: IMMUNE SYSTEM |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: HYDROLASE |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: HYDROLASE |
 |
Organism: Thaumatococcus daniellii
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: PLANT PROTEIN Ligands: TLA |
 |
Organism: Thaumatococcus daniellii
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: PLANT PROTEIN Ligands: TLA |

