Search Count: 17
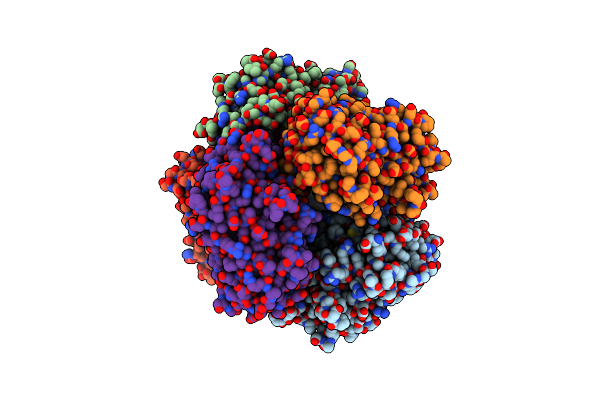 |
Organism: Streptomyces lividans 1326
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2025-05-28 Classification: OXIDOREDUCTASE Ligands: HEM, MG, O |
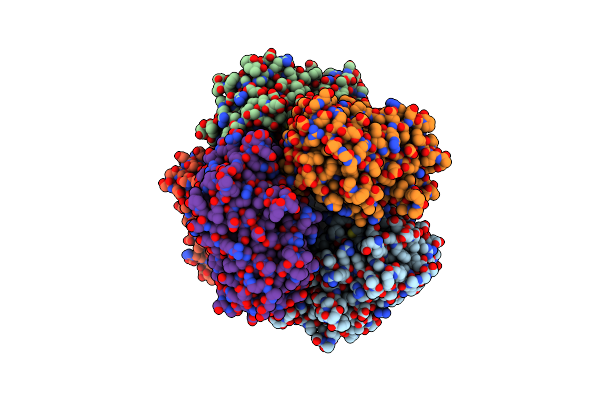 |
Organism: Streptomyces lividans 1326
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-05-28 Classification: OXIDOREDUCTASE Ligands: HEM, MG, O |
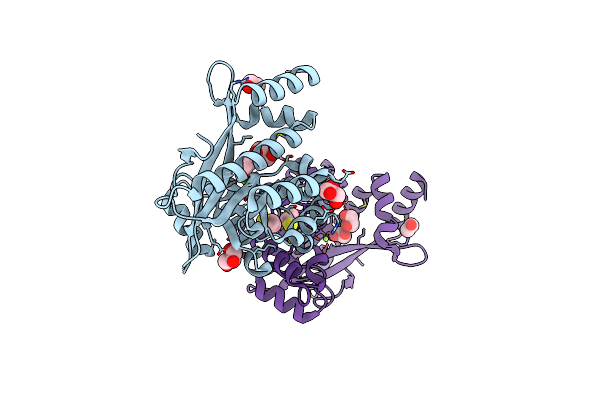 |
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2024-05-15 Classification: HYDROLASE Ligands: GOL, EDO, BME, MG |
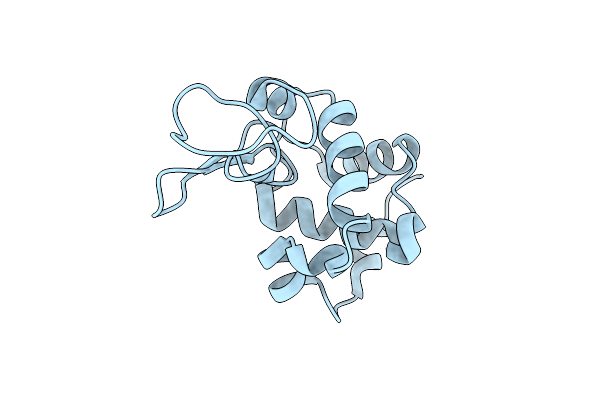 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-02-28 Classification: HYDROLASE Ligands: CL |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-02-28 Classification: HYDROLASE Ligands: CL |
 |
Ssx Structure Of Arabidopsis Thaliana Pdx1.3 Grown In Seeded Batch Conditions
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2024-02-28 Classification: LYASE Ligands: PO4 |
 |
Ssx Structure Of Arabidopsis Thaliana Pdx1.3 Grown In Microfluidic Droplets
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2024-02-28 Classification: LYASE Ligands: CIT, PO4 |
 |
Organism: Prochlorococcus marinus subsp. pastoris str. ccmp1986
Method: NEUTRON DIFFRACTION Resolution:2.10 Å Release Date: 2024-01-17 Classification: METAL BINDING PROTEIN Ligands: FE |
 |
Organism: Prochlorococcus marinus subsp. pastoris str. ccmp1986
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2023-08-30 Classification: METAL BINDING PROTEIN Ligands: FE |
 |
Organism: Prochlorococcus marinus subsp. pastoris str. ccmp1986
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2023-08-30 Classification: METAL BINDING PROTEIN Ligands: FE |
 |
Organism: Prochlorococcus marinus subsp. pastoris str. ccmp1986
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2023-08-30 Classification: METAL BINDING PROTEIN Ligands: FE2 |
 |
Organism: Prochlorococcus marinus subsp. pastoris str. ccmp1986
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2023-08-30 Classification: METAL BINDING PROTEIN Ligands: FE |
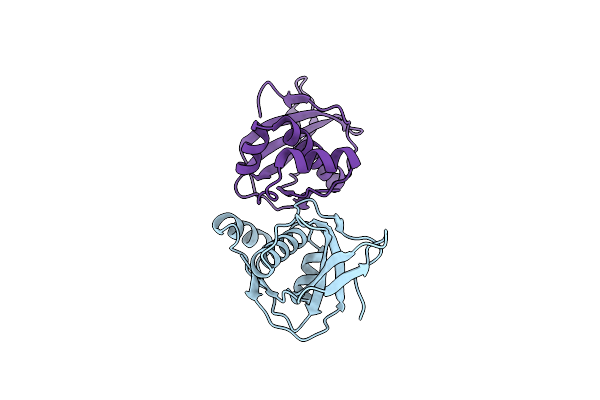 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.47 Å Release Date: 2023-08-16 Classification: PROTEIN BINDING |
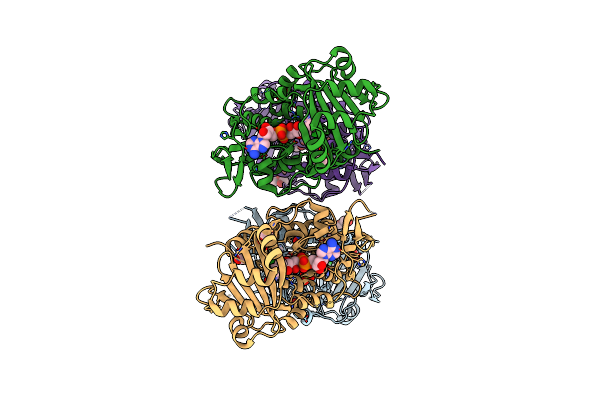 |
Organism: Stenotrophomonas maltophilia
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2023-07-12 Classification: OXIDOREDUCTASE Ligands: FAD, EDO, SO4, NO3, PEG, CL |
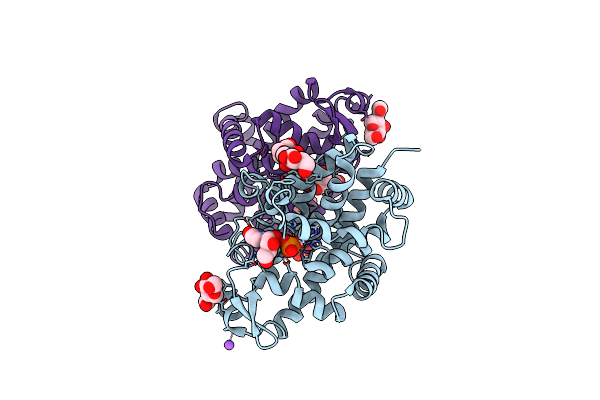 |
Organism: Aspergillus oryzae rib40
Method: X-RAY DIFFRACTION Resolution:1.06 Å Release Date: 2022-11-09 Classification: HYDROLASE Ligands: ZN, NAG, PO4, URI, NA, BTB |
 |
S1 Nuclease From Aspergillus Oryzae In Complex With Cytidine-5'-Monophosphate
Organism: Aspergillus oryzae rib40
Method: X-RAY DIFFRACTION Resolution:1.04 Å Release Date: 2022-11-09 Classification: HYDROLASE Ligands: ZN, NAG, PO4, C5P, BTB, CA |
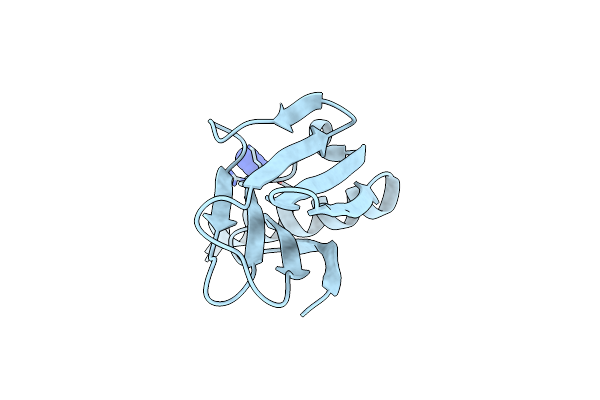 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2021-03-17 Classification: CONTRACTILE PROTEIN Ligands: AZI |

