Search Count: 33
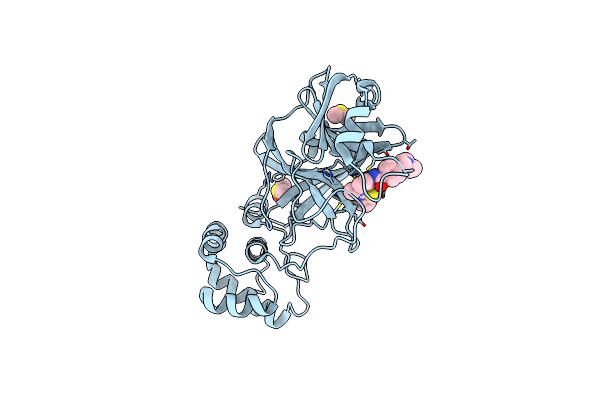 |
The Crystal Structure Of Sars-Cov-2 Omicron Mpro (P132H) In Complex With Demethylated Analog Of Masitinib
Organism: Severe acute respiratory syndrome coronavirus
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2022-02-16 Classification: HYDROLASE/INHIBITOR Ligands: XNJ, DMS |
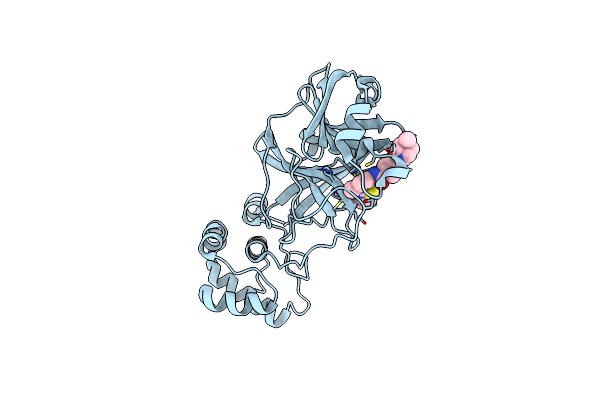 |
The Crystal Structure Of Sars-Cov-2 Omicron Mpro (P132H) In Complex With Masitinib
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2022-02-16 Classification: HYDROLASE/INHIBITOR Ligands: G65 |
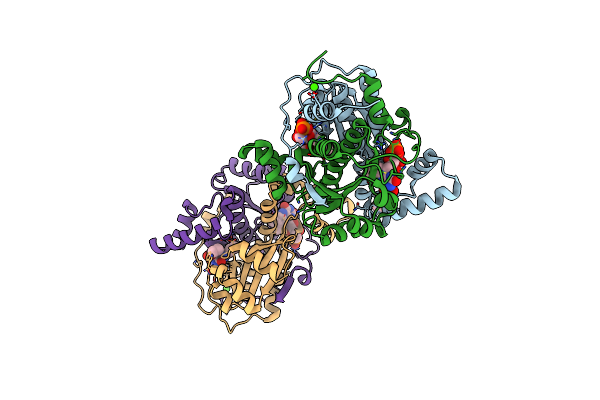 |
Crystal Structure Of Putative Nad(P)H-Flavin Oxidoreductase From Haemophilus Influenzae 86-028Np
Organism: Haemophilus influenzae (strain 86-028np)
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2021-10-06 Classification: OXIDOREDUCTASE Ligands: FMN, EDO, K, CL, CA |
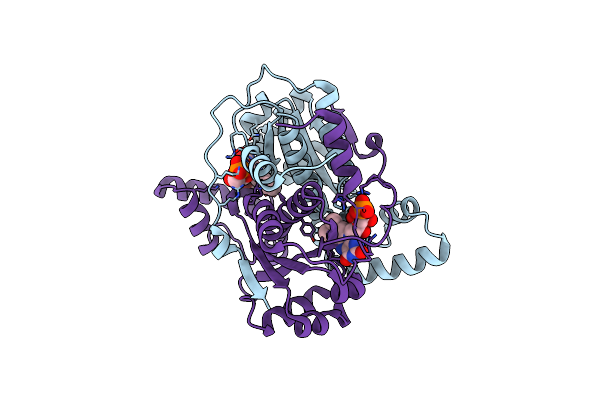 |
Crystal Structure Of Putative Nad(P)H-Flavin Oxidoreductase From Haemophilus Influenzae Rd Kw20
Organism: Haemophilus influenzae (strain atcc 51907 / dsm 11121 / kw20 / rd)
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2021-10-06 Classification: OXIDOREDUCTASE Ligands: FMN, CL, ACY |
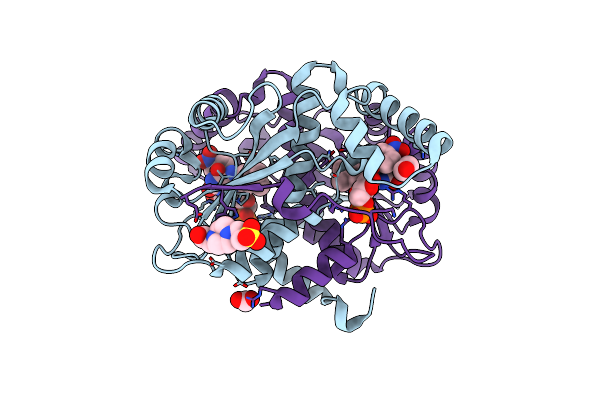 |
Crystal Structure Of Putative Nad(P)H-Flavin Oxidoreductase From Haemophilus Influenzae R2846 In Complex With 4-Nitrophenol
Organism: Haemophilus influenzae r2846
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2021-09-29 Classification: OXIDOREDUCTASE Ligands: NPO, EDO, FMT, FMN, EPE |
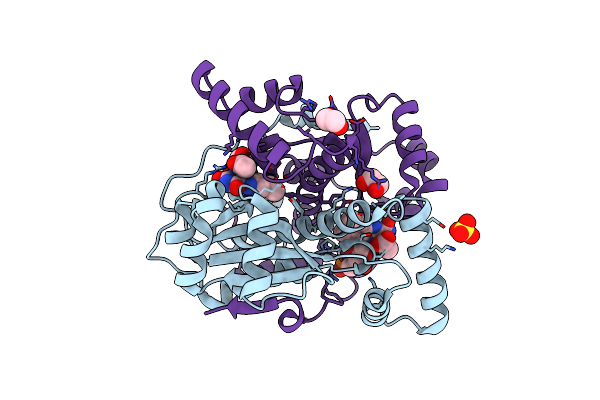 |
Crystal Structure Of Putative Nad(P)H-Flavin Oxidoreductase From Haemophilus Influenzae R2866
Organism: Haemophilus influenzae r2866
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2021-09-29 Classification: OXIDOREDUCTASE Ligands: FMN, EDO, SO4, ACY |
 |
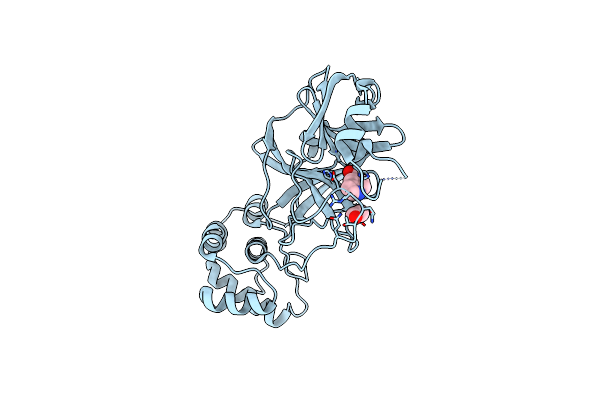
The Crystal Structure Of Sars-Cov-2 Main Protease In Complex With Demethylated Analog Of Masitinib
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2020-12-30 Classification: HYDROLASE Ligands: XNJ, DMS, GOL |
 |
The Crystal Structure Of Sars-Cov-2 Main Protease With The Formation Of Cys145-1H-Indole-5-Carboxylate
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2020-12-16 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: XC4, EDO |
 |
Crystal Structure Of Tryptophan Synthase From M. Tuberculosis - Aminoacrylate- And Gsk1-Bound Form
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Resolution:2.41 Å Release Date: 2020-09-30 Classification: LYASE/Lyase Inhibitor Ligands: FMT, MLI, PZJ, P1T, K, EDO, PEG, ACT, SO4, NA |
 |
The Crystal Structure Of Sars-Cov-2 Main Protease In Complex With Masitinib
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2020-09-09 Classification: HYDROLASE Ligands: G65, DMS, GOL |
 |
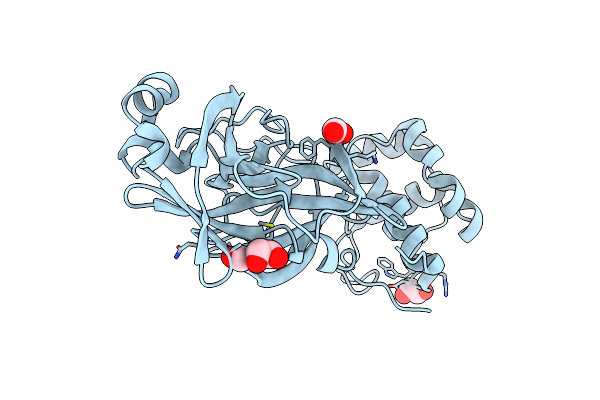
Crystal Structure Of Tryptophan Synthase From M. Tuberculosis - Aminoacrylate- And Gsk2-Bound Form
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2020-09-02 Classification: Lyase/Lyase Inhibitor Ligands: FMT, EDO, ACT, MLI, PZV, P1T, PEG, K, EPE, ALA, PGE, NA |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2020-07-29 Classification: HYDROLASE Ligands: EDO, FMT |
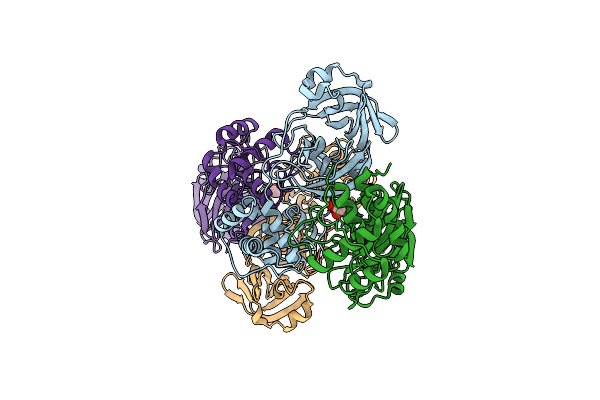 |
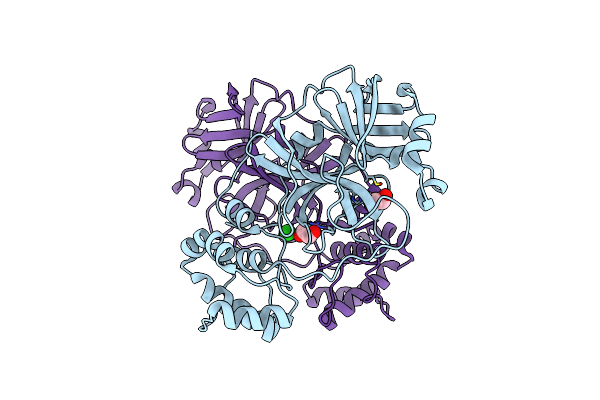
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2020-07-15 Classification: HYDROLASE Ligands: EDO |
 |
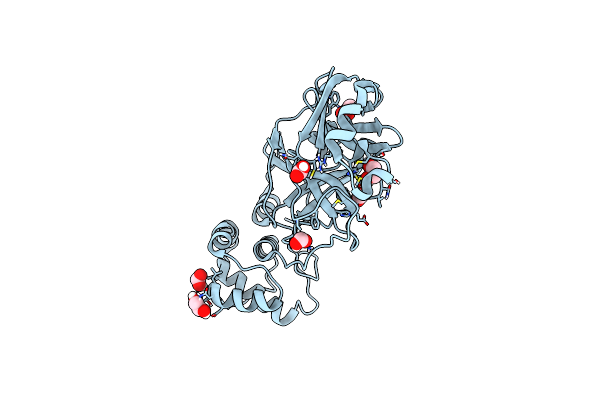
The Crystal Structure Of 3Cl Mainpro Of Sars-Cov-2 With Oxidized Cys145 (Sulfenic Acid Cysteine).
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-07-08 Classification: HYDROLASE Ligands: EDO, CL |
 |
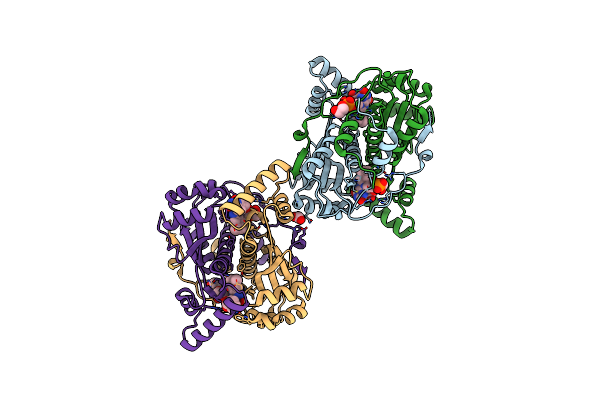
The 1.28A Crystal Structure Of 3Cl Mainpro Of Sars-Cov-2 With Oxidized C145 (Sulfinic Acid Cysteine)
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.28 Å Release Date: 2020-07-08 Classification: HYDROLASE Ligands: EDO, FMT, ACT |
 |
Crystal Structure Of Putative Nad(P)H-Flavin Oxidoreductase From Neisseria Meningitidis
Organism: Neisseria meningitidis serogroup a / serotype 4a (strain z2491)
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2020-05-13 Classification: OXIDOREDUCTASE Ligands: NIO, CL, FMN, EDO, FMT |
 |
Crystal Structure Of Adp Ribose Phosphatase Of Nsp3 From Sars-Cov-2 In Complex With Mes
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.07 Å Release Date: 2020-04-15 Classification: HYDROLASE Ligands: MES |
 |
Crystal Structure Of Adp Ribose Phosphatase Of Nsp3 From Sars-Cov-2 In The Apo Form
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2020-04-15 Classification: HYDROLASE Ligands: CL |
 |
Crystal Structure Of The Methyltransferase-Stimulatory Factor Complex Of Nsp16 And Nsp10 From Sars Cov-2.
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-03-25 Classification: VIRAL PROTEIN Ligands: CL, EDO, SAM, ZN |
 |
Crystal Structure Of Adp Ribose Phosphatase Of Nsp3 From Sars Cov-2 In Complex With Amp
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2020-03-25 Classification: VIRAL PROTEIN, HYDROLASE Ligands: AMP, MES |

