Search Count: 27
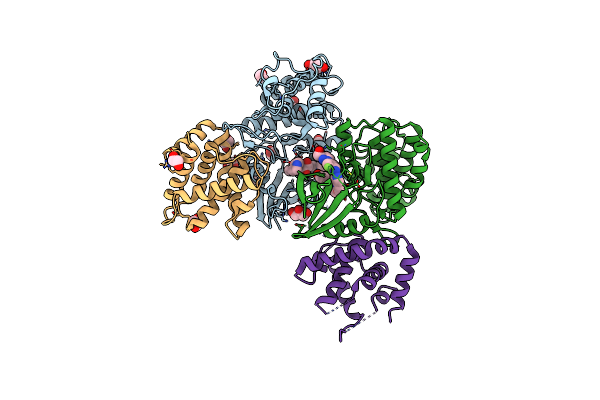 |
The Structure Of Cyclin-Dependent Kinase 5 (Cdk5) In Complex With P25 And Compound 1
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2022-03-09 Classification: TRANSFERASE Ligands: 65L, EDO, PEG, CL, GOL |
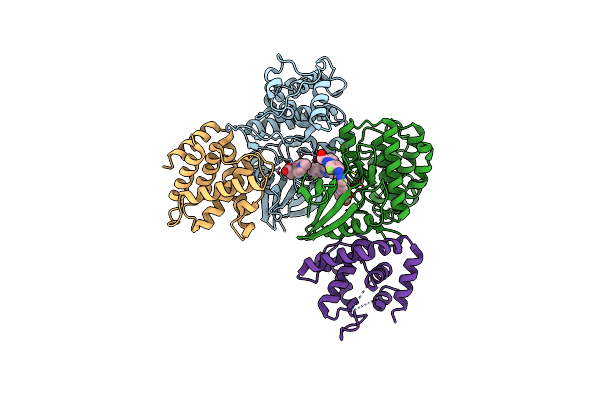 |
The Structure Of Cyclin-Dependent Kinase 5 (Cdk5) In Complex With P25 And Compound 7
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.91 Å Release Date: 2022-03-09 Classification: TRANSFERASE Ligands: 64V |
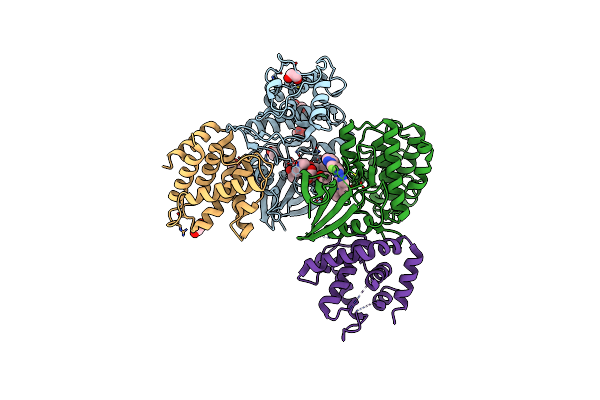 |
The Structure Of Cyclin-Dependent Kinase 5 (Cdk5) In Complex With P25 And Compound 13
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2022-03-09 Classification: TRANSFERASE Ligands: 63I, PGE, EDO |
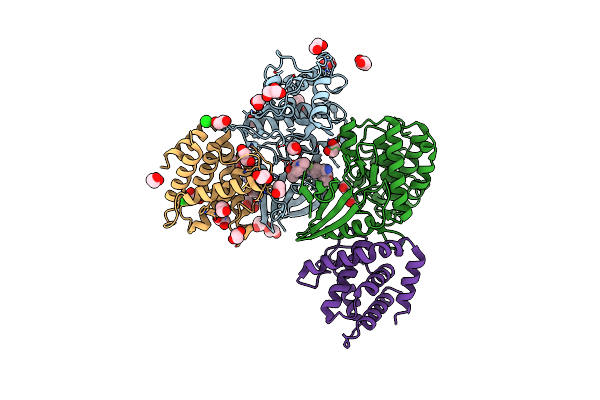 |
The Structure Of Cyclin-Dependent Kinase 5 (Cdk5) In Complex With P25 And Compound 24
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.05 Å Release Date: 2022-03-09 Classification: TRANSFERASE Ligands: 61U, GOL, EDO, CL |
 |
The Structure Of Cyclin-Dependent Kinase 2 (Cdk2) In Complex With Compound 1
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.53 Å Release Date: 2022-03-09 Classification: TRANSFERASE Ligands: 65L |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2021-11-03 Classification: TRANSFERASE/INHIBITOR Ligands: 3I3 |
 |
Organism: Danio rerio
Method: ELECTRON MICROSCOPY Release Date: 2020-12-09 Classification: TRANSPORT PROTEIN Ligands: SJQ, CA, 44E |
 |
Organism: Danio rerio
Method: ELECTRON MICROSCOPY Release Date: 2020-12-09 Classification: TRANSPORT PROTEIN Ligands: LPP, CA |
 |
Organism: Danio rerio
Method: ELECTRON MICROSCOPY Release Date: 2020-12-09 Classification: TRANSPORT PROTEIN Ligands: S9Q, CA, 44E |
 |
Organism: Danio rerio
Method: ELECTRON MICROSCOPY Release Date: 2020-12-09 Classification: TRANSPORT PROTEIN Ligands: SKQ, CA, LPP |
 |
Organism: Danio rerio, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2020-12-09 Classification: TRANSPORT PROTEIN Ligands: CA, 44E |
 |
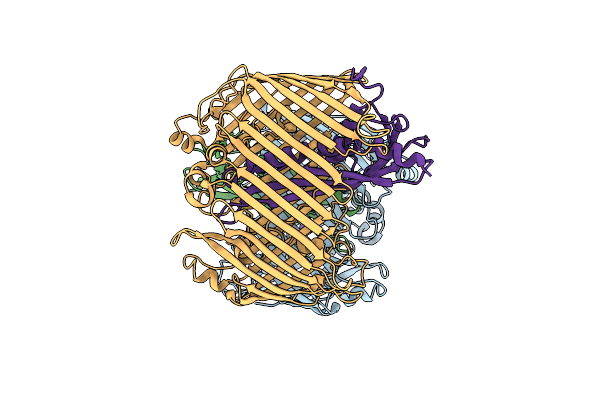
Crystal Structure Of The Outer Membrane Lipopolysaccharide Transport Protein Lpte (Rlpb) From Escherichia Coli In The Tetragonal Crystal Form
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2015-11-11 Classification: LIPID BINDING PROTEIN Ligands: FMT |
 |
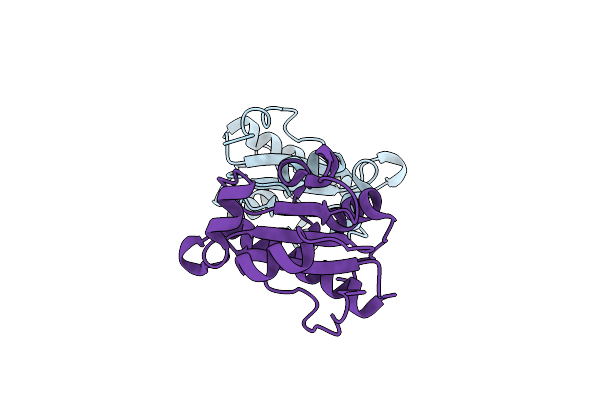
Crystal Structure Of The Lipopolysaccharide Assembly Complex Lptd-Lpte From The Escherichia Coli Outer Membrane
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.36 Å Release Date: 2015-08-26 Classification: TRANSPORT PROTEIN Ligands: CL, NA |
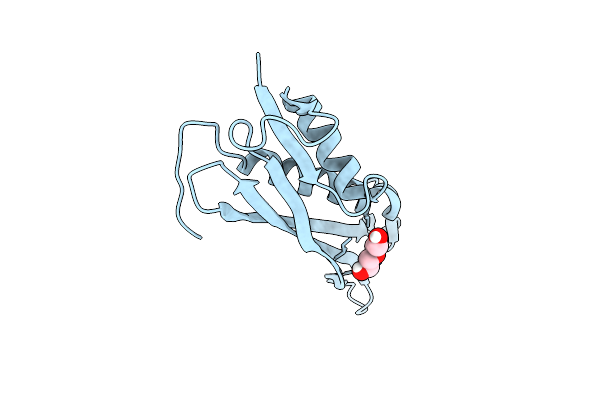 |
Crystal Structure Of The Outer Membrane Lipopolysaccharide Transport Protein Lpte (Rlpb)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.34 Å Release Date: 2014-07-16 Classification: LIPID BINDING PROTEIN Ligands: PEG |
 |
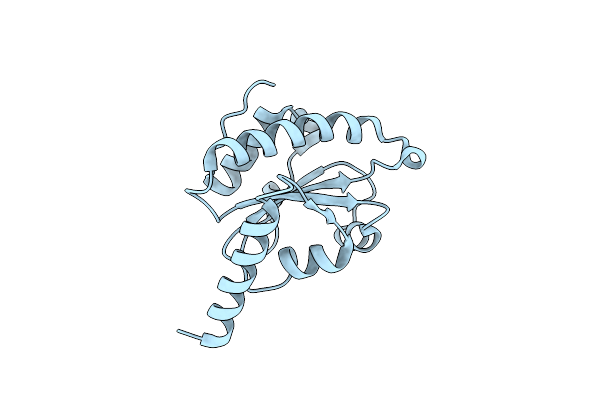
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2014-03-26 Classification: OXIDOREDUCTASE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2014-03-26 Classification: OXIDOREDUCTASE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2014-03-26 Classification: OXIDOREDUCTASE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2014-03-26 Classification: OXIDOREDUCTASE |
 |
Apo Form Of Bacterial Arylsulfate Sulfotransferase (Asst) H436N Mutant With Mpo In The Active Site
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2014-03-26 Classification: TRANSFERASE Ligands: MPO, SO4 |
 |
Bacterial Arylsulfate Sulfotransferase (Asst) H436N Mutant With 4-Nitrophenyl Sulfate (Pns) In The Active Site
Organism: Escherichia coli cft073
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2014-03-26 Classification: TRANSFERASE Ligands: 4NS, SO4 |

