Search Count: 16
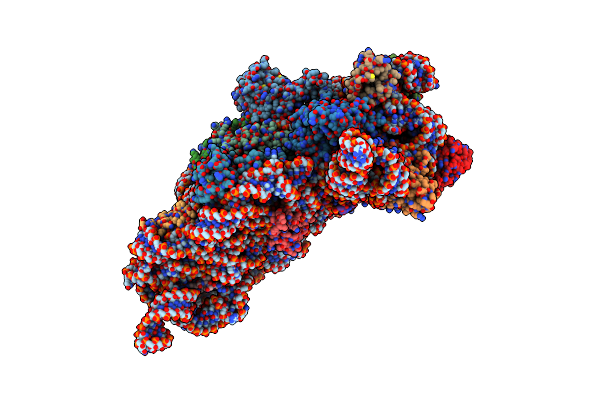 |
Structure Of The Thermus Thermophilus 30S Ribosomal Subunit Complexed With A Human Anti-Codon Stem Loop (Hasl) Of Transfer Rna Lysine 3 (Trnalys3) Bound To An Mrna With An Aaa-Codon In The A-Site And Paromomycin
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:3.11 Å Release Date: 2012-01-25 Classification: Ribosome/Antibiotic Ligands: MG, PAR, ZN |
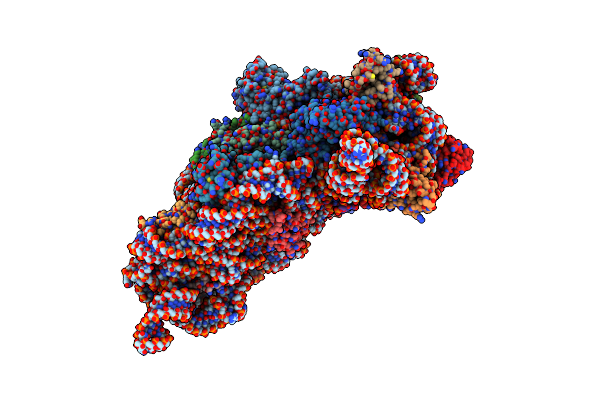 |
Structure Of The Thermus Thermophilus 30S Ribosomal Subunit Complexed With A Human Anti-Codon Stem Loop (Hasl) Of Transfer Rna Lysine 3 (Trnalys3) Bound To An Mrna With An Aag-Codon In The A-Site And Paromomycin
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2012-01-25 Classification: Ribosome/Antibiotic Ligands: MG, PAR, ZN |
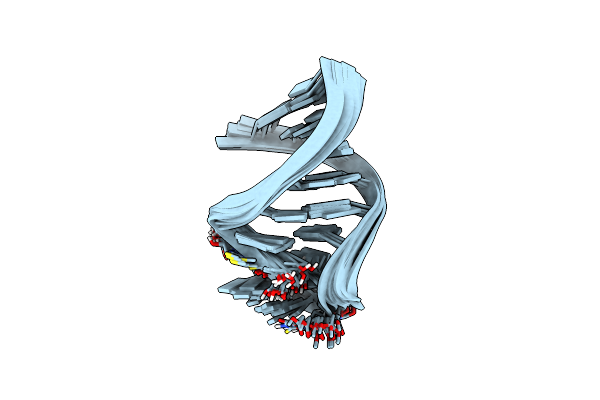 |
Solution Structure Of The Human Anti-Codon Stem And Loop(Hasl) Of Transfer Rna Lysine 3 (Trnalys3)
|
 |
 |
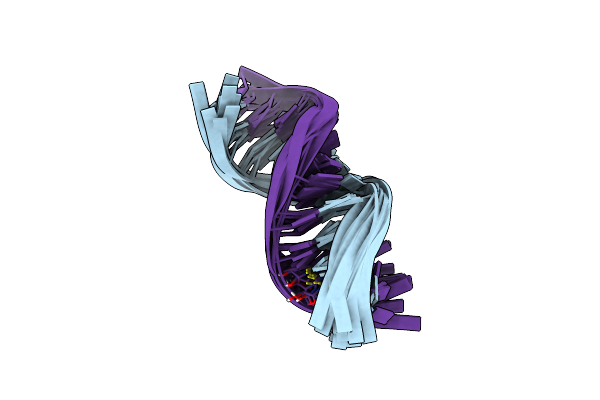
Nmr Solution Structure Of The Anticodon Of E.Coli Trna-Val3 With 1 Modification (M6A37)
|
 |
Nmr Solution Structure Of The Anticodon Of E.Coli Trna-Val3 With No Modifications
|
 |
Nmr Solution Structure Of The Anticodon Of E. Coli Trna-Val3 With 2 Modifications (Cmo5U34 M6A37)
|
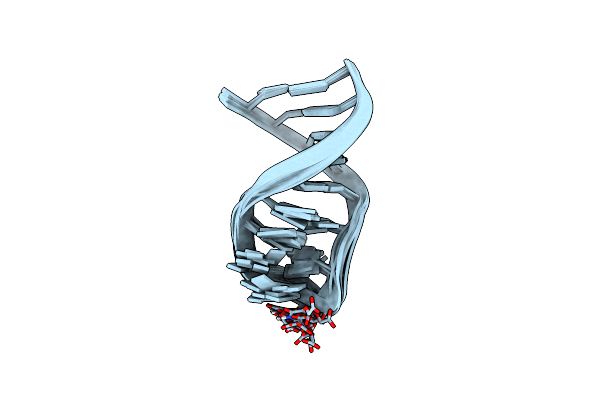 |
Nmr Solution Structure Of The Anticodon Of E. Coli Trna-Val3 With 1 Modification (Cmo5U34)
|
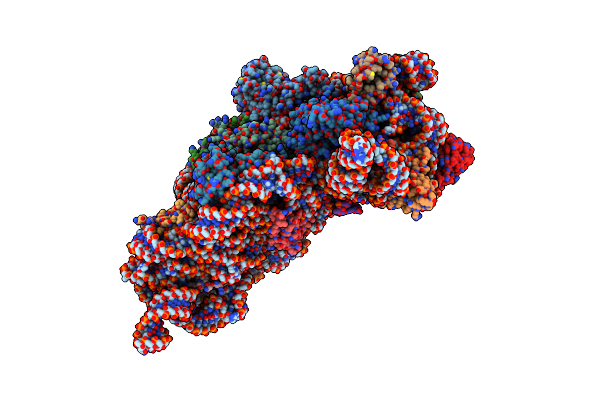 |
Structure Of The Thermus Thermophilus 30S Ribosomal Subunit Complexed With A Valine-Asl With Cmo5U In Position 34 Bound To An Mrna With A Gua-Codon In The A-Site And Paromomycin.
Organism: Synthetic construct, Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2007-05-22 Classification: RIBOSOME Ligands: PAR, MG, K, ZN |
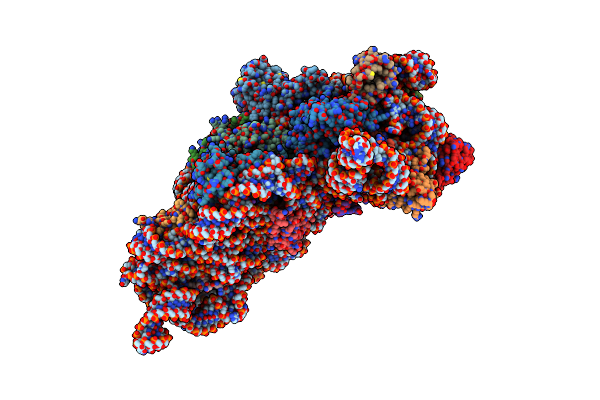 |
Structure Of The Thermus Thermophilus 30S Ribosomal Subunit Complexed With A Valine-Asl With Cmo5U In Position 34 Bound To An Mrna With A Gug-Codon In The A-Site And Paromomycin.
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2007-05-15 Classification: RIBOSOME Ligands: PAR, MG, K, ZN |
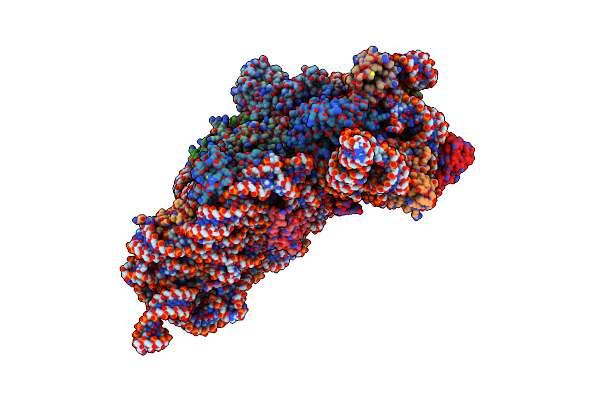 |
Structure Of The Thermus Thermophilus 30S Ribosomal Subunit Complexed With A Valine-Asl With Cmo5U In Position 34 Bound To An Mrna With A Guc-Codon In The A-Site And Paromomycin.
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Release Date: 2007-05-15 Classification: RIBOSOME Ligands: PAR, MG, K, ZN |
 |
Structure Of The Thermus Thermophilus 30S Ribosomal Subunit Complexed With A Valine-Asl With Cmo5U In Position 34 Bound To An Mrna With A Guu-Codon In The A-Site And Paromomycin.
Organism: Synthetic construct, Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2007-05-15 Classification: RIBOSOME Ligands: PAR, MG, K, ZN |
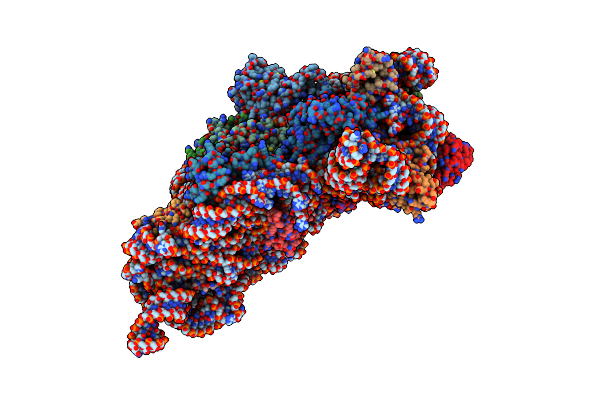 |
Crystal Structure Of Mnm5U34T6A37-Trnalysuuu Complexed With Aag-Mrna In The Decoding Center
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:3.25 Å Release Date: 2004-12-14 Classification: RIBOSOME Ligands: PAR, MG, ZN |
 |
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2004-12-14 Classification: RIBOSOME Ligands: PAR, MG, ZN |
 |
Nmr Solution Structure Of The Anticodon Of Trna(Lys3) With T6A Modification At Position 37
|
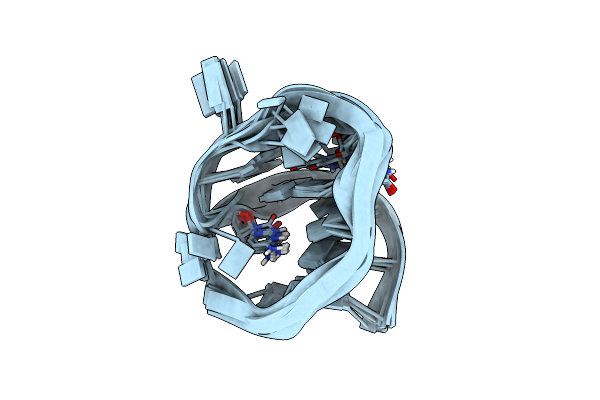 |
Solution Nmr Structure Of An Analog Of The Yeast Trna Phe T Stem Loop Containing Ribothymidine At Its Naturally Occurring Position
|

