Search Count: 6
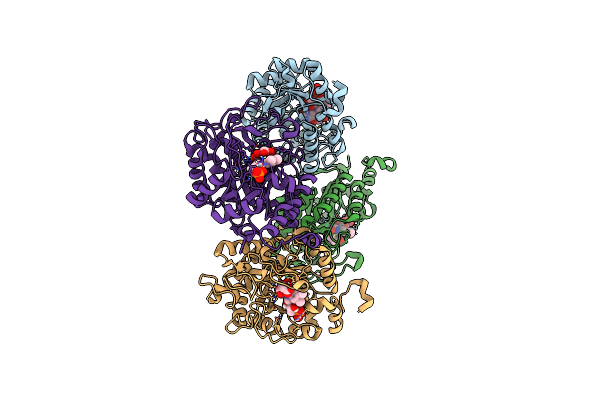 |
Succinate Bound Crystal Structure Of Thermus Scotoductus Sa-01 Ene-Reductase
Organism: Thermus scotoductus sa-01
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2024-07-03 Classification: OXIDOREDUCTASE Ligands: SIN, FMN |
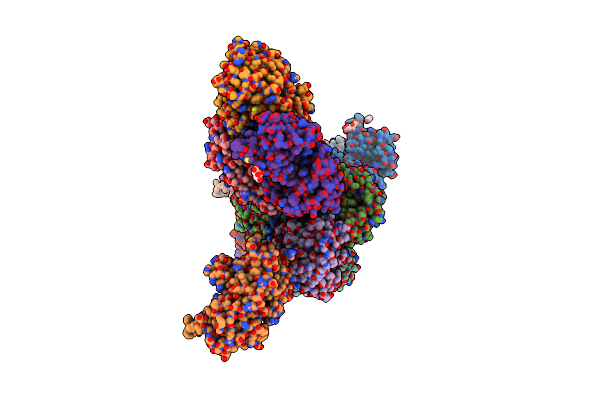 |
Rhodococcus Ruber Alcohol Dehydrogenase Nadh Biomimetic Complex - Compound 4B
Organism: Rhodococcus ruber
Method: X-RAY DIFFRACTION Resolution:2.99 Å Release Date: 2024-07-03 Classification: OXIDOREDUCTASE Ligands: ZN, W46, CIT, IPA |
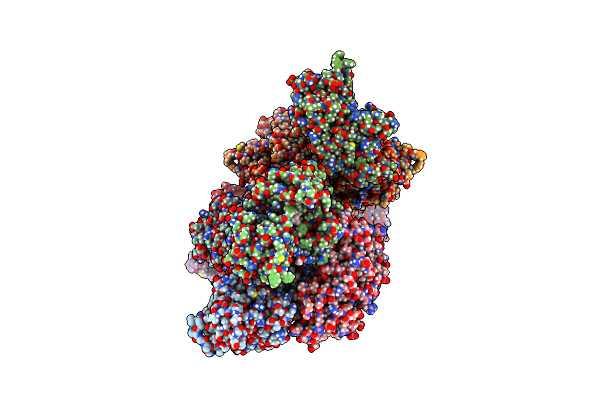 |
Rhodococcus Ruber Alcohol Dehydrogenase Nadh Biomimetic Complex - Compound 1A
Organism: Rhodococcus ruber
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-07-03 Classification: OXIDOREDUCTASE Ligands: IPA, ZN, NA, W3O, CIT |
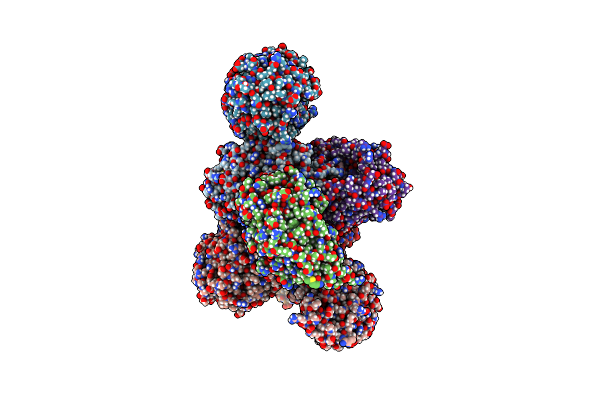 |
Organism: Thermus scotoductus sa-01
Method: X-RAY DIFFRACTION Resolution:2.76 Å Release Date: 2024-07-03 Classification: FLAVOPROTEIN Ligands: FMN, W3X, IPA, NA, CL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-07-12 Classification: HYDROLASE Ligands: ACT, SO4, NH4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2023-07-12 Classification: HYDROLASE Ligands: ACT, SO4 |

