Search Count: 17
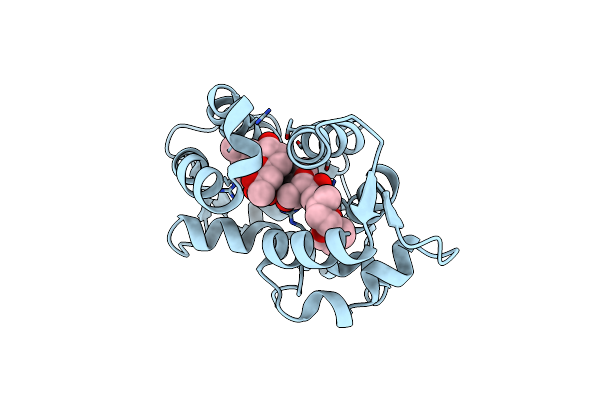 |
Organism: Halichondria okadai
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2015-05-27 Classification: TOXIN/TOXIN INHIBITOR Ligands: OKA |
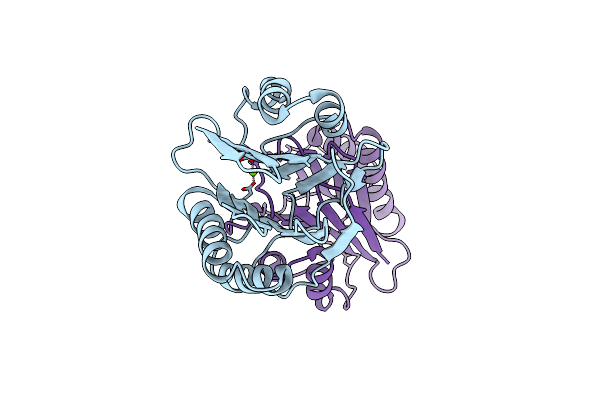 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2014-01-22 Classification: HYDROLASE Ligands: MG |
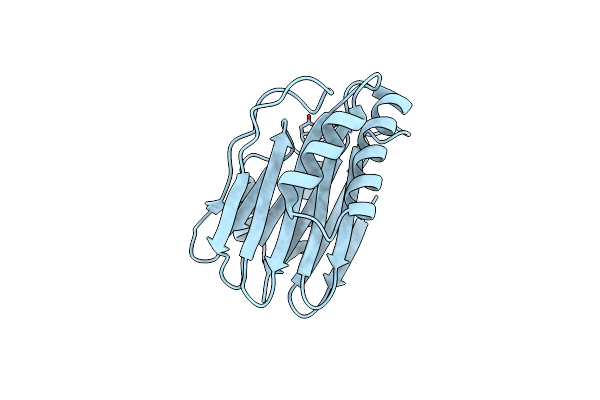 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.42 Å Release Date: 2014-01-22 Classification: HYDROLASE Ligands: MG |
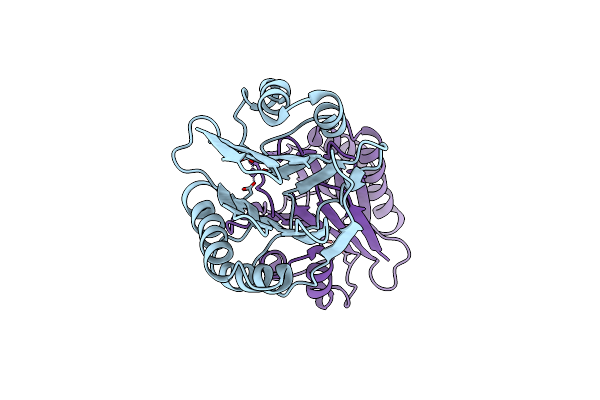 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.06 Å Release Date: 2014-01-22 Classification: HYDROLASE Ligands: MN |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.22 Å Release Date: 2014-01-22 Classification: HYDROLASE Ligands: MN |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2014-01-22 Classification: HYDROLASE Ligands: MN, PEG |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2014-01-22 Classification: HYDROLASE Ligands: CO |
 |
Structural Basis Of Glycine Amide On Suppression Of Protein Aggregation By High Resolution X-Ray Analysis
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.05 Å Release Date: 2011-02-16 Classification: HYDROLASE Ligands: GM1, CL, NA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2011-02-02 Classification: OXYGEN TRANSPORT Ligands: HEM, CMO |
 |
Crystal Structure Of 11,11'-Dichlorochromopyrrolic Acid Bound Cytochrome P450 Stap (Cyp245A1)
Organism: Streptomyces sp. tp-a0274
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2009-05-12 Classification: OXIDOREDUCTASE Ligands: HEM, 2CC |
 |
Crystal Structure Of Myoglobin Reconstituted With 6-Methyl-6-Depropionatehemin
Organism: Physeter catodon
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2007-08-14 Classification: OXYGEN STORAGE/TRANSPORT Ligands: SO4, 6HE |
 |
Crystal Structure Of Myoglobin Reconstituted With 7-Methyl-7-Depropionatehemin
Organism: Physeter catodon
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2007-08-14 Classification: OXYGEN STORAGE/TRANSPORT Ligands: SO4, 7HE |
 |
Organism: Streptomyces sp. tp-a0274
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2007-07-03 Classification: OXIDOREDUCTASE Ligands: HEM, IMD, EDO |
 |
Crystal Structure Of Chromopyrrolic Acid Bound Cytochrome P450 Stap (Cyp245A1)
Organism: Streptomyces sp. tp-a0274
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2007-07-03 Classification: OXIDOREDUCTASE Ligands: HEM, CRR, EDO |
 |
Organism: Physeter catodon
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2006-10-31 Classification: OXYGEN STORAGE/TRANSPORT Ligands: HME, IMD |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2006-05-23 Classification: OXYGEN STORAGE/TRANSPORT Ligands: HEM, ACY |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2004-06-08 Classification: OXYGEN STORAGE/TRANSPORT Ligands: HEM |

