Search Count: 35
 |
Crystal Structure Of Calcium/Calmodulin-Dependent Protein Kinase Type Ii Subunit Delta Complexed With Ribociclib
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2025-05-07 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: 6ZZ |
 |
Crystal Structure Of Actin Capping Protein In Complex With A Fragment Of Legionella Pneumophila Ravb
Organism: Gallus gallus, Legionella pneumophila subsp. pneumophila (strain philadelphia 1 / atcc 33152 / dsm 7513)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-05-07 Classification: PROTEIN BINDING Ligands: FMT |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-03-12 Classification: VIRAL PROTEIN/RNA/INHIBITOR Ligands: ZN, A1AWQ, MN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2024-04-03 Classification: PROTEIN BINDING |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2024-04-03 Classification: PROTEIN BINDING |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2023-05-31 Classification: Cell Cycle, Transferase Ligands: EDO, 1QK, 7TW |
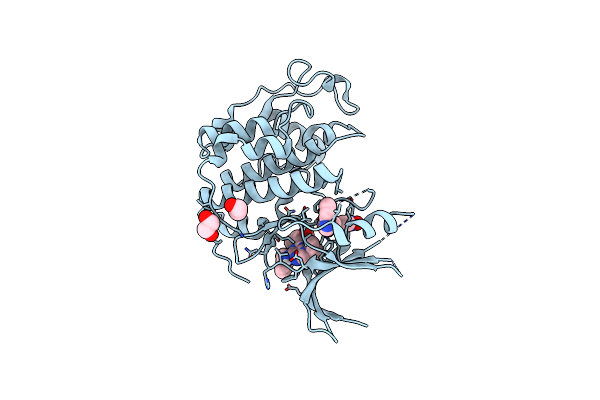 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2023-05-31 Classification: Cell Cycle, Transferase Ligands: EDO, RRC, 7TW |
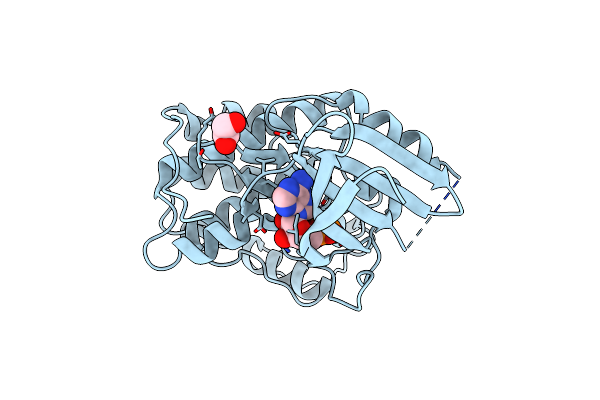 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2023-05-31 Classification: Cell Cycle, Transferase Ligands: EDO, ATP, MG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-09-28 Classification: CELL CYCLE Ligands: 8IL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2022-08-24 Classification: CELL CYCLE Ligands: EDO, 7TW |
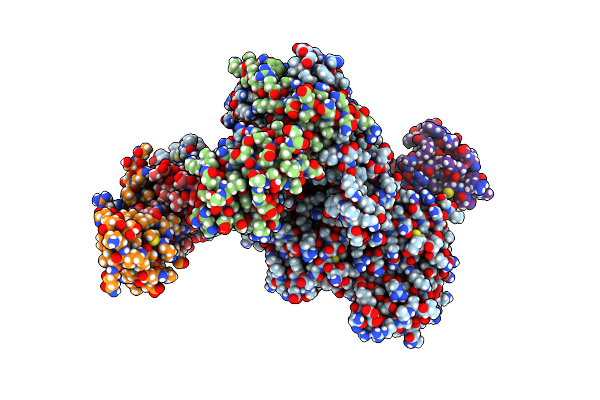 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2022-03-16 Classification: VIRAL PROTEIN Ligands: ZN, MN, POP |
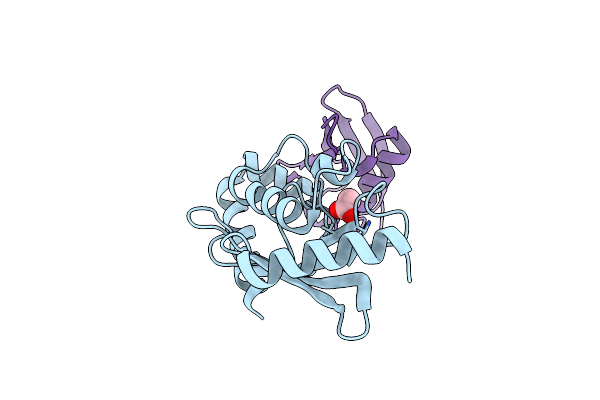 |
Organism: Saccharomyces cerevisiae, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2015-09-02 Classification: NUCLEAR PROTEIN Ligands: ACT |
 |
Crystal Structure Of Staphylococcal Nuclease Variant Nviaga/E122G At Cryogenic Temperature
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2012-05-23 Classification: HYDROLASE Ligands: PO4 |
 |
Crystal Structure Of Staphylococcal Nuclease Variant Nviaga V99G At Cryogenic Temperature
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2012-04-18 Classification: HYDROLASE Ligands: PO4, MPD |
 |
Crystal Structure Of Staphylococcal Nuclease Variant Nviaga/N100G At Cryogenic Temperature
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2011-11-23 Classification: HYDROLASE Ligands: PO4 |
 |
Crystal Structure Of Staphylococcal Nuclease Variant Delta+Nviagla V23E At Cryogenic Temperature
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2011-09-14 Classification: HYDROLASE Ligands: THP, MRD |
 |
Crystal Structure Of Staphylococcal Nuclease Variant Nviaga/M98G At Cryogenic Temperature
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2011-08-03 Classification: HYDROLASE |
 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2011-06-08 Classification: RNA Binding Protein/RNA |
 |
Organism: Escherichia coli
Method: SOLUTION NMR Release Date: 2011-03-23 Classification: DE NOVO PROTEIN |
 |
Crystal Structure Of Staphylococcal Nuclease Variant Delta+Phs D21N At Cryogenic Temperature
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2011-02-16 Classification: HYDROLASE Ligands: THP, PO4 |

