Search Count: 35
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2018-05-30 Classification: TRANSFERASE Ligands: EDO, PEG, DXE, M2M, B3P, 6L7 |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2017-08-16 Classification: TRANSFERASE Ligands: 6YB, EDO, DXE, PEG, P33 |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2017-06-21 Classification: TRANSFERASE Ligands: GJV, DXE, EDO, PG4, PEG |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2017-06-21 Classification: TRANSFERASE Ligands: 6ZH, EDO, PEG, PG4, PE8, DXE, M2M |
 |
Crystal Structure Of Mouse Carm1 In Complex With Sah At 1.8 Angstroms Resolution
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2017-03-29 Classification: TRANSFERASE Ligands: SAH, EDO, PEG, PGE, PE8, DXE |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-03-29 Classification: TRANSFERASE Ligands: 6CX, EDO, PEG, M2M, PG4 |
 |
Crystal Structure Of Mouse Carm1 In Complex With Sinefungin At 2.0 Angstroms Resolution
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2017-03-15 Classification: TRANSFERASE Ligands: PG4, EDO, PEG, DXE, SFG |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2017-03-15 Classification: TRANSFERASE Ligands: EDO, PEG, DSH |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.71 Å Release Date: 2017-03-15 Classification: TRANSFERASE Ligands: 6D1, EDO, SAH |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2017-03-15 Classification: TRANSFERASE Ligands: 6D3, EDO, PEG, DXE, PG4, M2M |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2017-03-15 Classification: TRANSFERASE Ligands: 6D5, EDO, DXE, PEG, M2M, PG4, NA |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2017-03-15 Classification: TRANSFERASE Ligands: SAO, EDO, 6D0, DXE, M2M |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2017-03-15 Classification: TRANSFERASE Ligands: EDO, PEG, M2M, SAO, 6CZ |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2017-03-15 Classification: TRANSFERASE Ligands: 6D2, EDO, PGE, DXE, PEG, PE8 |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.22 Å Release Date: 2017-03-15 Classification: TRANSFERASE Ligands: 6D4, EDO, PEG, PG4, M2M, DXE, SAO |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.42 Å Release Date: 2017-03-15 Classification: TRANSFERASE Ligands: DXE, EDO, PG6, PEG, 6D6, M2M |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2017-03-15 Classification: TRANSFERASE Ligands: 765, EDO, PEG, DXE, M2M, PG4 |
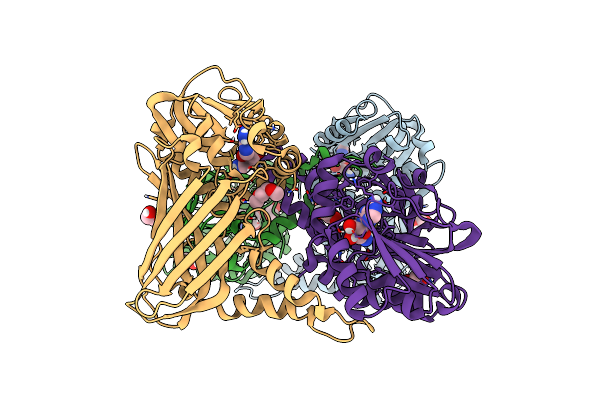 |
Crystal Structure Of Mouse Carm1 In Complex With Inhibitor Sa0920 (5'-Amino-5'-Deoxyadenosine)
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.74 Å Release Date: 2017-03-15 Classification: TRANSFERASE Ligands: 5N5, EDO, DXE |
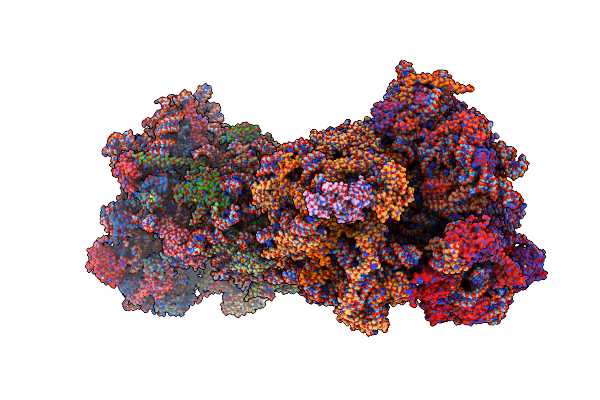 |
Coping With Proline Stalling: Structural Basis Of Hypusine-Induced Protein Synthesis By The Eukaryotic Ribosome
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c), Saccharomyces cerevisiae s288c
Method: X-RAY DIFFRACTION Resolution:3.45 Å Release Date: 2017-01-25 Classification: RIBOSOME Ligands: MG, OHX, ZN, SPS, PRO |
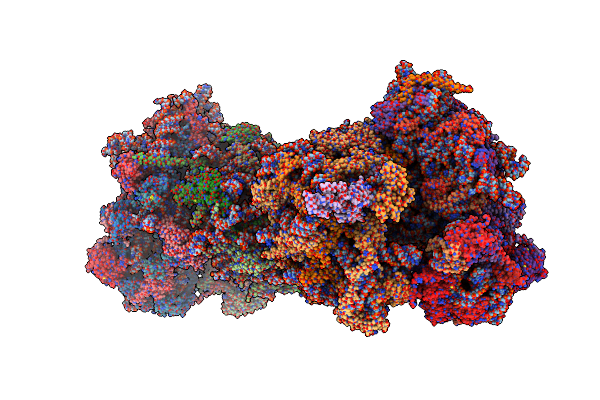 |
Crystal Structure Of The S.Cerevisiae 80S Ribosome In Complex With The A-Site Bound Aminoacyl-Trna Analog Acca-Pro
Organism: Homo sapiens, Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2017-01-18 Classification: RIBOSOME Ligands: OHX, MG, ZN, PHE, LEU, SPS, 8AN |

