Search Count: 8
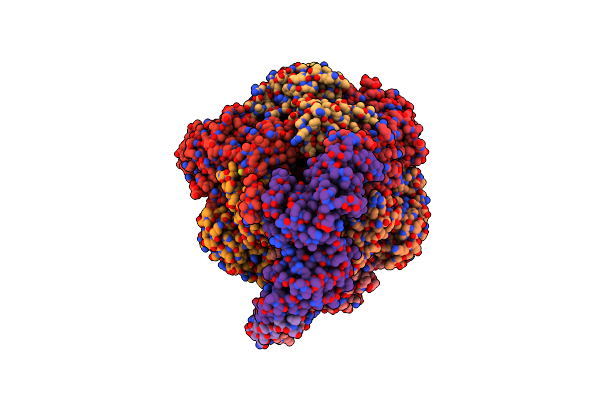 |
Cryo-Em Structure Of Bedaquiline-Free Mycobacterium Smegmatis Atp Synthase Rotational State 1
Organism: Mycolicibacterium smegmatis
Method: ELECTRON MICROSCOPY Release Date: 2020-08-19 Classification: TRANSLOCASE Ligands: ATP, MG, PO4, ADP |
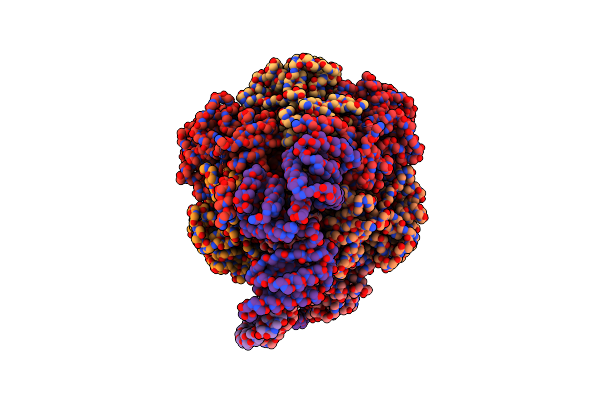 |
Cryo-Em Structure Of Bedaquiline-Free Mycobacterium Smegmatis Atp Synthase Rotational State 2 (Backbone Model)
Organism: Mycolicibacterium smegmatis
Method: ELECTRON MICROSCOPY Release Date: 2020-08-19 Classification: TRANSLOCASE |
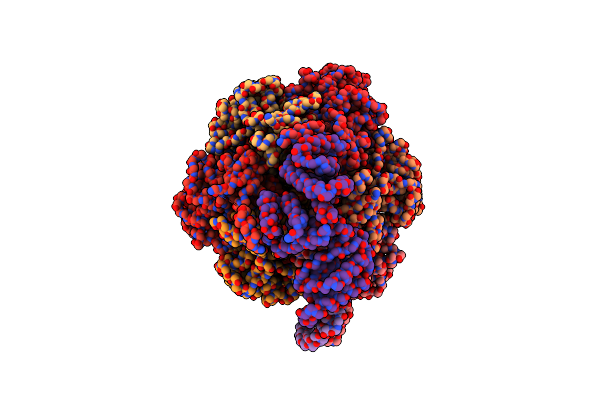 |
Cryo-Em Structure Of Bedaquiline-Free Mycobacterium Smegmatis Atp Synthase Rotational State 3 (Backbone Model)
Organism: Mycolicibacterium smegmatis
Method: ELECTRON MICROSCOPY Release Date: 2020-08-19 Classification: TRANSLOCASE |
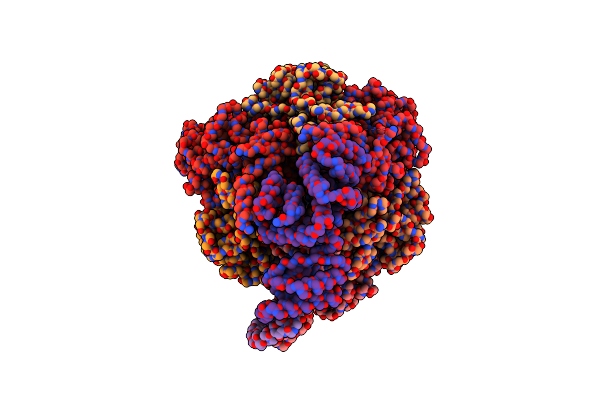 |
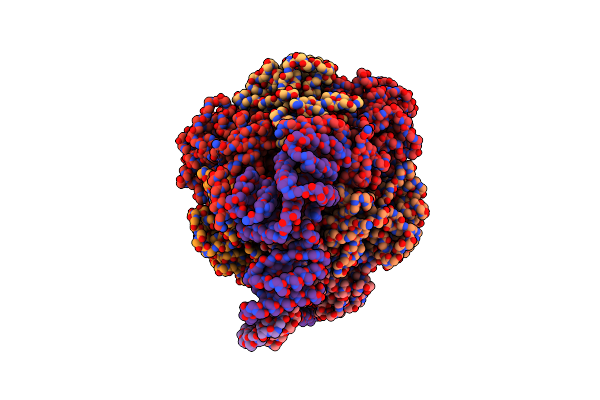
Cryo-Em Structure Of Bedaquiline-Saturated Mycobacterium Smegmatis Atp Synthase Rotational State 1 (Backbone Model)
Organism: Mycolicibacterium smegmatis
Method: ELECTRON MICROSCOPY Release Date: 2020-08-19 Classification: HYDROLASE Ligands: BQ1 |
 |
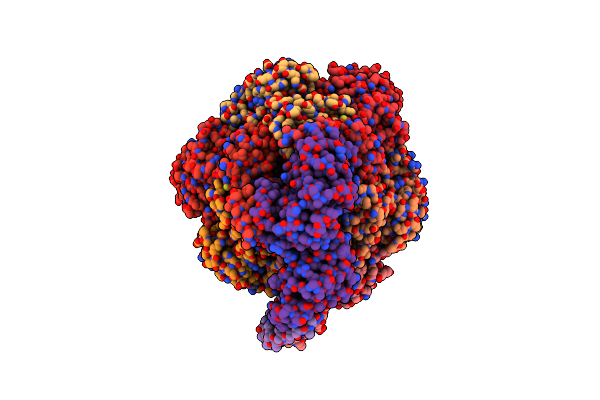
Cryo-Em Structure Of Bedaquiline-Saturated Mycobacterium Smegmatis Atp Synthase Rotational State 2 (Backbone Model)
Organism: Mycolicibacterium smegmatis
Method: ELECTRON MICROSCOPY Release Date: 2020-08-19 Classification: HYDROLASE Ligands: BQ1 |
 |
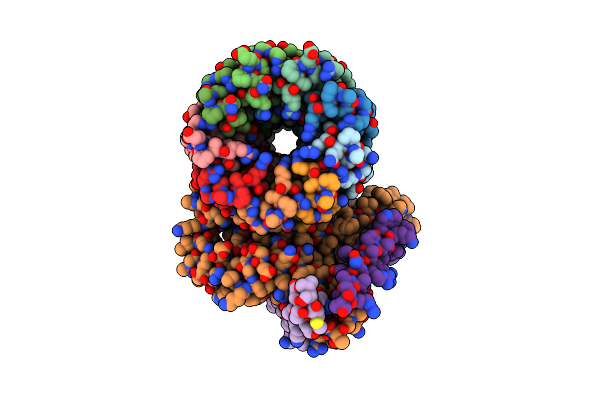
Cryo-Em Structure Of Bedaquiline-Saturated Mycobacterium Smegmatis Atp Synthase Rotational State 3
Organism: Mycolicibacterium smegmatis
Method: ELECTRON MICROSCOPY Release Date: 2020-08-19 Classification: HYDROLASE Ligands: ATP, MG, ADP, PO4, BQ1 |
 |
Cryo-Em Structure Of Bedaquiline-Free Mycobacterium Smegmatis Atp Synthase Fo Region
Organism: Mycolicibacterium smegmatis
Method: ELECTRON MICROSCOPY Release Date: 2020-08-19 Classification: HYDROLASE |
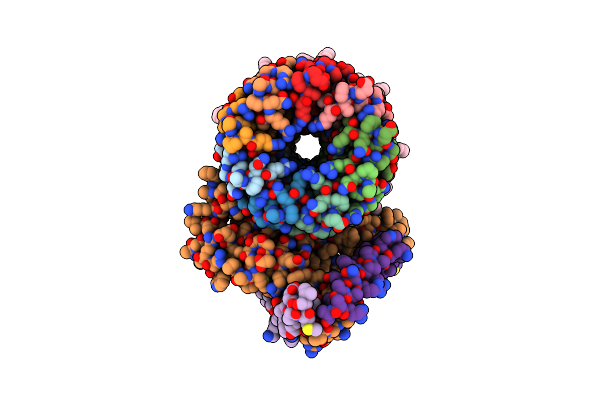 |
Cryo-Em Structure Of Bedaquiline-Saturated Mycobacterium Smegmatis Atp Synthase Fo Region
Organism: Mycolicibacterium smegmatis
Method: ELECTRON MICROSCOPY Release Date: 2020-08-19 Classification: HYDROLASE Ligands: BQ1 |

