Search Count: 71
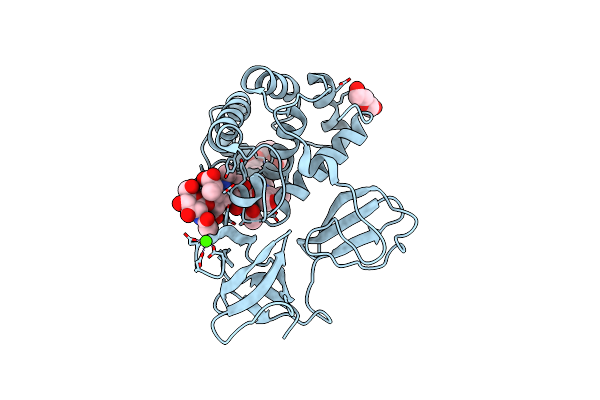 |
Crystal Structure Of Catalytic Domain Of Lytb (E585Q) From Streptococcus Pneumoniae In Complex With Nag-Nam-Nag-Nam Tetrasaccharide
Organism: Streptococcus pneumoniae (strain atcc baa-255 / r6)
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2022-09-21 Classification: HYDROLASE Ligands: 1PE, PEG, CA |
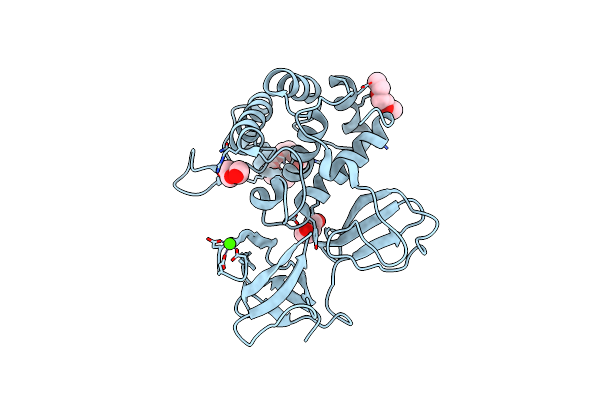 |
Crystal Structure Of Catalytic Domain In Open Conformation Of Lytb From Streptococcus Pneumoniae
Organism: Streptococcus pneumoniae (strain atcc baa-255 / r6)
Method: X-RAY DIFFRACTION Resolution:1.43 Å Release Date: 2022-09-07 Classification: HYDROLASE Ligands: 1PE, PGE, PEG, CA |
 |
Crystal Structure Of Catalytic Domain In Closed Conformation Of Lytb (E585Q)From Streptococcus Pneumoniae
Organism: Streptococcus pneumoniae (strain atcc baa-255 / r6)
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2022-09-07 Classification: HYDROLASE Ligands: 1PE, PGE, PEG, ACT, CA |
 |
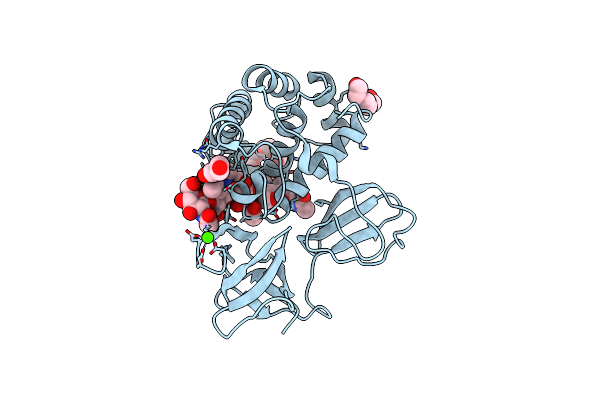
Crystal Structure Of Catalytic Domain Of Lytb From Streptococcus Pneumoniae In Complex With Nag-Nag-Nag-Nag Tetrasaccharide
Organism: Streptococcus pneumoniae (strain atcc baa-255 / r6)
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2022-09-07 Classification: HYDROLASE Ligands: 1PE, PEG, CA |
 |
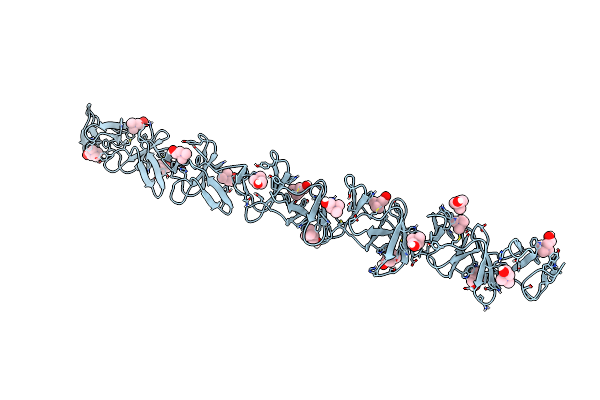
Crystal Structure Of Catalytic Domain Of Lytb (E585Q) From Streptococcus Pneumoniae In Complex With Nag-Nam-Nag-Nam-Nag Peptidolycan Analogue
Organism: Streptococcus pneumoniae (strain atcc baa-255 / r6)
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2022-09-07 Classification: HYDROLASE Ligands: 1PE, PGE, PEG, CA |
 |
Crystal Structure Of Choline-Binding Module Of Lytb From Streptococcus Pneumoniae
Organism: Streptococcus pneumoniae (strain atcc baa-255 / r6)
Method: X-RAY DIFFRACTION Resolution:2.98 Å Release Date: 2022-09-07 Classification: HYDROLASE Ligands: CHT |
 |
Crystal Structure Of Catalytic Domain In Closed Conformation Of Lytb From Streptococcus Pneumoniae
Organism: Streptococcus pneumoniae r6
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2022-09-07 Classification: HYDROLASE Ligands: 1PE, PEG, CA |
 |
Crystal Structure Of Choline-Binding Module (R1-R9) Of Lytb From Streptococcus Pneumoniae
Organism: Streptococcus pneumoniae (strain atcc baa-255 / r6)
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2022-09-07 Classification: HYDROLASE Ligands: CHT, ZN, PGE |
 |
Crystal Structure Of Penicillin-Binding Protein 1 (Pbp1) From Staphylococcus Aureus
Organism: Staphylococcus aureus subsp. aureus col
Method: X-RAY DIFFRACTION Resolution:3.03 Å Release Date: 2021-11-03 Classification: HYDROLASE Ligands: EPE, CD, CL |
 |
Crystal Structure Of Penicillin-Binding Protein 1 (Pbp1) From Staphylococcus Aureus In Complex With Piperacillin
Organism: Staphylococcus aureus subsp. aureus col
Method: X-RAY DIFFRACTION Resolution:3.03 Å Release Date: 2021-11-03 Classification: HYDROLASE Ligands: YPP |
 |
Crystal Structure Of Penicillin-Binding Protein 1 (Pbp1) From Staphylococcus Aureus In Complex With Penicillin G
Organism: Staphylococcus aureus subsp. aureus col
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2021-11-03 Classification: HYDROLASE Ligands: CIT, SO4, PNM |
 |
Crystal Structure Of Pasta Domains Of The Penicillin-Binding Protein 1 (Pbp1) From Staphylococcus Aureus
Organism: Staphylococcus aureus subsp. aureus col
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2021-11-03 Classification: HYDROLASE Ligands: CL |
 |
Crystal Structure Of Penicillin-Binding Protein 1 (Pbp1) From Staphylococcus Aureus In Complex With Pentaglycine
Organism: Staphylococcus aureus (strain col), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.36 Å Release Date: 2021-11-03 Classification: HYDROLASE Ligands: CD, CL |
 |
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-04-15 Classification: HYDROLASE Ligands: SO4, MG |
 |
The Crystal Structure Of Glycoside Hydrolase Bglx From P. Aeruginosa In Complex With 1-Deoxynojirimycin
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-04-15 Classification: HYDROLASE Ligands: PGE, NOJ, MG |
 |
The Crystal Structure The Glycoside Hydrolase Bglx Inactive Mutant D286N From P. Aeruginosa In Complex With Two Glucose Molecules
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2020-04-15 Classification: HYDROLASE Ligands: BGC, MG |
 |
The Crystal Structure Of Glycoside Hydrolase Bglx Inactive Mutant D286N From P. Aeruginosa In Complex With Glucose
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2020-04-15 Classification: HYDROLASE Ligands: BGC, MG |
 |
The Crystal Structure Of Glycoside Hydrolase Bglx Inactive Mutant D286N From P. Aeruginosa In Complex With Cellobiose
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2020-04-15 Classification: HYDROLASE Ligands: PEG, PGE, MG |
 |
The Crystal Structure Of Glycoside Hydrolase Bglx Inactive Mutant D286N From P. Aeruginosa In Complex With Lactose
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2020-04-15 Classification: HYDROLASE Ligands: PEG, MG, PGE |
 |
The Crystal Structure Of Glycoside Hydrolase Bglx Inactive Mutant From P. Aeruginosa In Complex With Laminaritriose
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2020-04-15 Classification: HYDROLASE Ligands: MG |

