Search Count: 12
 |
The Crystal Structure Of Tfua Involved In Peptide Backbone Thioamidation From Methanosarcina Acetivorans
Organism: Methanosarcina acetivorans
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2021-03-17 Classification: BIOSYNTHETIC PROTEIN |
 |
Structure Of Ycao Enzyme From Methanocaldococcus Jannaschii In Complex With Atp
Organism: Methanocaldococcus jannaschii (strain atcc 43067 / dsm 2661 / jal-1 / jcm 10045 / nbrc 100440)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2020-01-08 Classification: BIOSYNTHETIC PROTEIN Ligands: ATP |
 |
Structure Of Ycao Enzyme From Methanocaldococcus Jannaschii In Complex With Peptide
Organism: Methanocaldococcus jannaschii (strain atcc 43067 / dsm 2661 / jal-1 / jcm 10045 / nbrc 100440), Methanocaldococcus jannaschii
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2019-11-06 Classification: BIOSYNTHETIC PROTEIN Ligands: ATP, MG |
 |
Organism: Deinococcus radiodurans (strain atcc 13939 / dsm 20539 / jcm 16871 / lmg 4051 / nbrc 15346 / ncimb 9279 / r1 / vkm b-1422)
Method: X-RAY DIFFRACTION Resolution:2.33 Å Release Date: 2019-04-03 Classification: LYASE Ligands: ACT, 3HB |
 |
The Structure Of Ycao From Methanopyrus Kandleri Bound With Amppcp And Mg2+
Organism: Methanopyrus kandleri (strain av19 / dsm 6324 / jcm 9639 / nbrc 100938)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2018-03-21 Classification: BIOSYNTHETIC PROTEIN Ligands: ACP, MG |
 |
The Structure Of Ycao From Methanopyrus Kandleri Bound With Amppcp And Mg2+
Organism: Methanopyrus kandleri (strain av19 / dsm 6324 / jcm 9639 / nbrc 100938)
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2018-03-21 Classification: BIOSYNTHETIC PROTEIN Ligands: ADP, MG, OXM |
 |
Crystal Structure Of Pseudouridine Monophosphate Glycosidase Complexed With Sulfate
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2012-10-31 Classification: HYDROLASE Ligands: SO4, MN |
 |
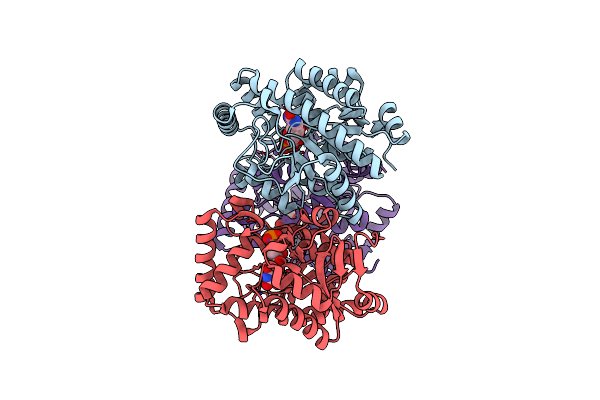
Crystal Structure Of Pseudouridine Monophosphate Glycosidase/Linear R5P Adduct
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2012-10-31 Classification: HYDROLASE Ligands: R5P, MN |
 |
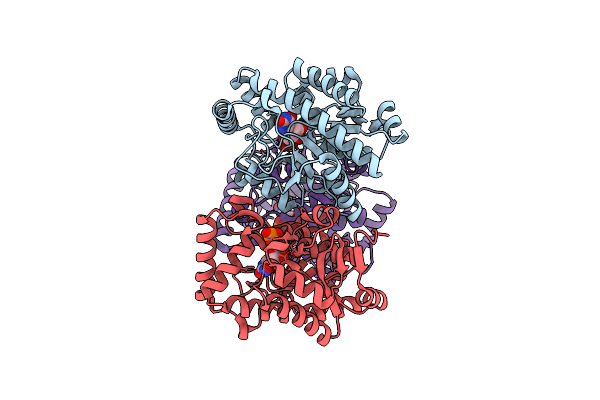
Crystal Structure Of Pseudouridine Monophosphate Glycosidase/Linear Pseudouridine 5'-Phosphate Adduct
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.54 Å Release Date: 2012-10-31 Classification: HYDROLASE Ligands: MN, KPS |
 |
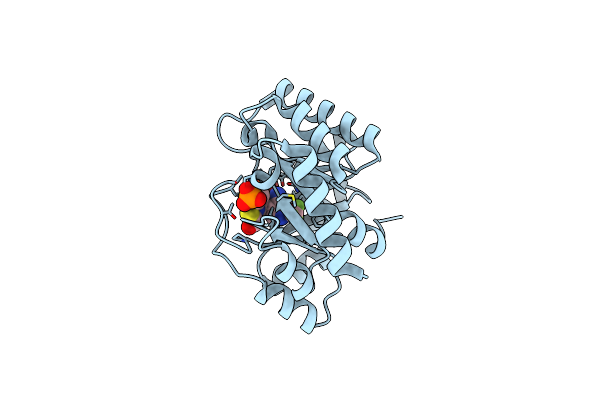
Crystal Structure Of Pseudouridine Monophosphate Glycosidase Complexed With Pseudouridine 5'-Phosphate
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2012-10-31 Classification: HYDROLASE Ligands: PSU, MN |
 |
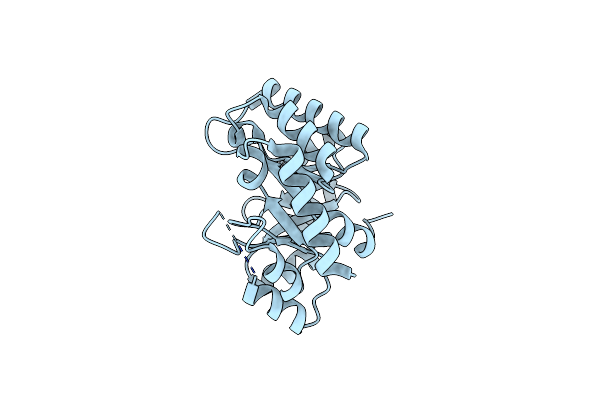
Crystal Structure Of Bacillus Subtilis Thiamin Phosphate Synthase Complexed With A Carboxylated Thiazole Phosphate
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2011-07-27 Classification: TRANSFERASE Ligands: 3NM, IFP, POP |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2011-07-27 Classification: TRANSFERASE |

