Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
Organism: Pseudomonas putida kt2440
Method: X-RAY DIFFRACTION Release Date: 2024-03-20 Classification: TRANSCRIPTION Ligands: SEY, PO4, CL |
 |
Organism: Pseudomonas putida kt2440
Method: X-RAY DIFFRACTION Release Date: 2024-03-20 Classification: TRANSCRIPTION Ligands: SIN, EDO, PO4, CL |
 |
Organism: Clostridium carboxidivorans p7
Method: X-RAY DIFFRACTION Resolution:2.38 Å Release Date: 2024-03-13 Classification: TRANSPORT PROTEIN Ligands: CL |
 |
Crystal Structure Of Sulfonamide Resistance Enzyme Sul1 In Complex With 6-Hydroxymethylpterin
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:2.32 Å Release Date: 2023-05-03 Classification: TRANSFERASE Ligands: HHR, GOL, SO4, CL |
 |
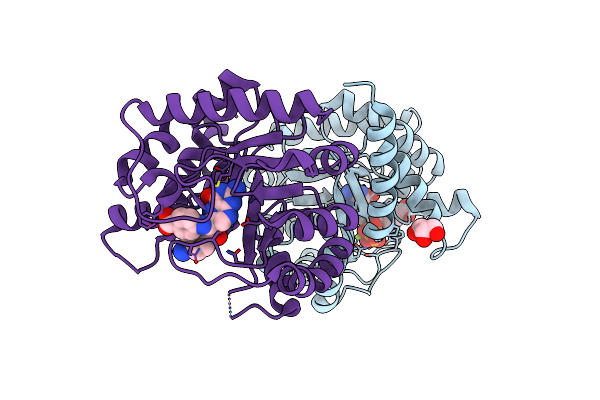
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2023-05-03 Classification: TRANSFERASE Ligands: SO4, CL, GOL, PEG |
 |
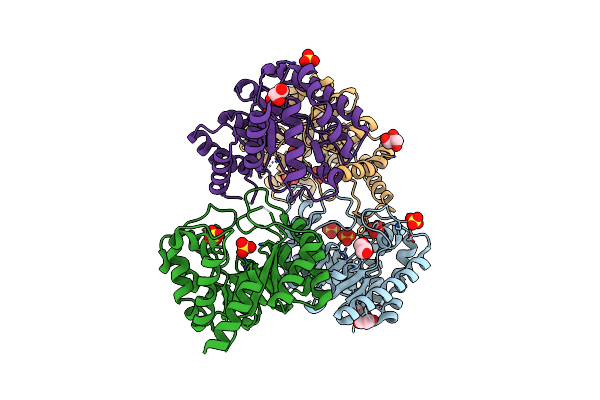
Crystal Structure Of Sulfonamide Resistance Enzyme Sul2 In Complex With 7,8-Dihydropteroate, Magnesium, And Pyrophosphate
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2023-05-03 Classification: TRANSFERASE Ligands: 78H, POP, MG, GOL, CL, PAB |
 |
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:2.79 Å Release Date: 2023-05-03 Classification: TRANSFERASE Ligands: GOL, CL, SO4 |
 |
Crystal Structure Of Sulfonamide Resistance Enzyme Sul3 In Complex With 6-Hydroxymethylpterin
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:2.42 Å Release Date: 2023-05-03 Classification: TRANSFERASE Ligands: HHR |
 |
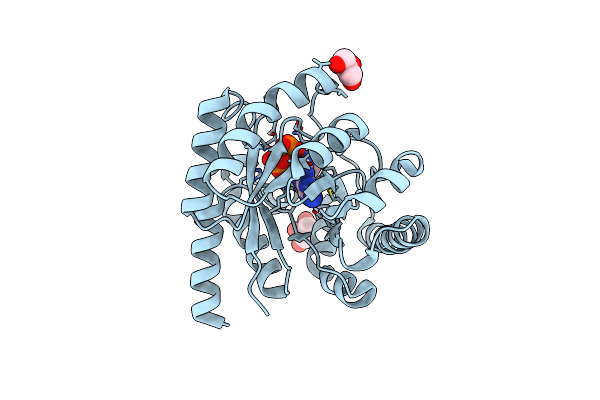
Crystal Structure Of Adaptive Laboratory Evolved Sulfonamide-Resistant Dihydropteroate Synthase (Dhps) From Escherichia Coli In Complex With 6-Hydroxymethylpterin
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.73 Å Release Date: 2023-05-03 Classification: TRANSFERASE Ligands: HHR |
 |
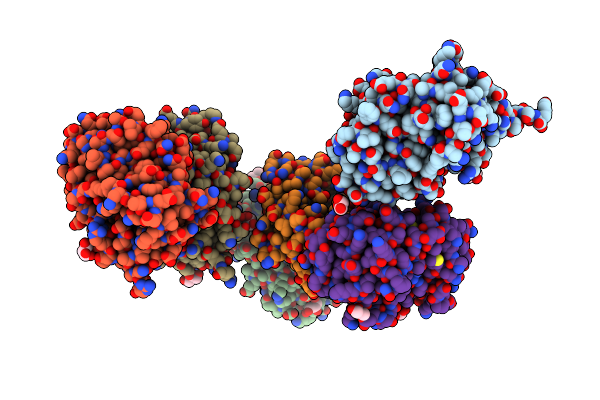
Crystal Structure Of Sulfonamide Resistance Enzyme Sul3 In Complex With Reaction Intermediate
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:2.06 Å Release Date: 2023-05-03 Classification: TRANSFERASE Ligands: XHP, PAB, GOL, CL, MG, POP |
 |
Crystal Structure Of Bh1352 2-Deoxyribose-5-Phosphate From Bacillus Halodurans, K184L Mutant
Organism: Bacillus halodurans c-125
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 2019-10-23 Classification: LYASE Ligands: GOL |
 |
Crystal Structure Of Bh1352 2-Deoxyribose-5-Phosphate From Bacillus Halodurans
Organism: Bacillus halodurans (strain atcc baa-125 / dsm 18197 / ferm 7344 / jcm 9153 / c-125)
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2019-10-16 Classification: TRANSFERASE Ligands: GOL, TRS |
 |
X-Ray Crystal Structure Of A Novel Aldo-Keto Reductases For The Biocatalytic Conversion Of 3-Hydroxybutanal To 1,3-Butanediol
Organism: Salmonella typhimurium (strain lt2 / sgsc1412 / atcc 700720)
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2017-02-15 Classification: TRANSLATION Ligands: SO4, CL, NDP |
 |
Crystal Structure Of A Putative Succinate-Semialdehyde Dehydrogenase From Salmonella Typhimurium Lt2 With Bound Nad
Organism: Salmonella typhimurium
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2008-11-04 Classification: OXIDOREDUCTASE Ligands: NAD |
 |
Structure Of Idp01002, A Putative Oxidoreductase From And Essential Gene Of Salmonella Typhimurium
Organism: Salmonella enterica subsp. enterica serovar typhimurium
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2008-11-04 Classification: OXIDOREDUCTASE Ligands: ZN, NA, CL, CAC, EDO |
 |
Crystal Structure Of A Putative Succinate-Semialdehyde Dehydrogenase From Salmonella Typhimurium Lt2
Organism: Salmonella typhimurium
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2008-11-04 Classification: OXIDOREDUCTASE |