Search Count: 47
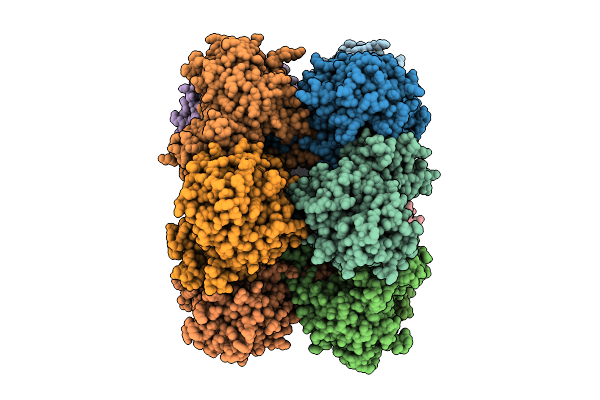 |
Organism: Azotobacter vinelandii
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: LIGASE |
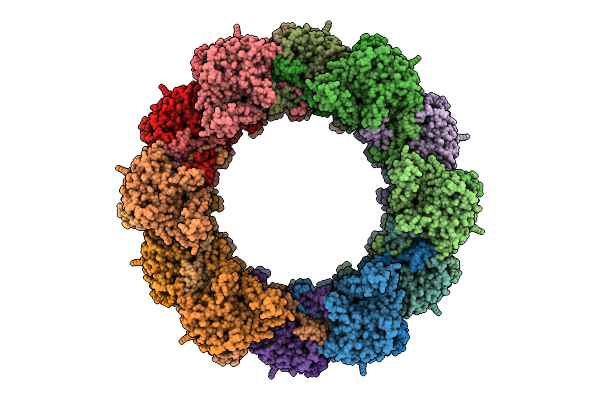 |
Azotobacter Vinelandii Extended Type Vi Secretion System Sheath Tube Complex
Organism: Azotobacter vinelandii
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: TRANSPORT PROTEIN |
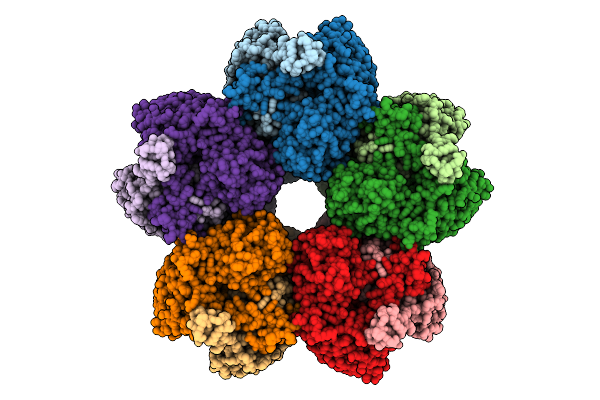 |
Organism: Azotobacter vinelandii
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: ISOMERASE |
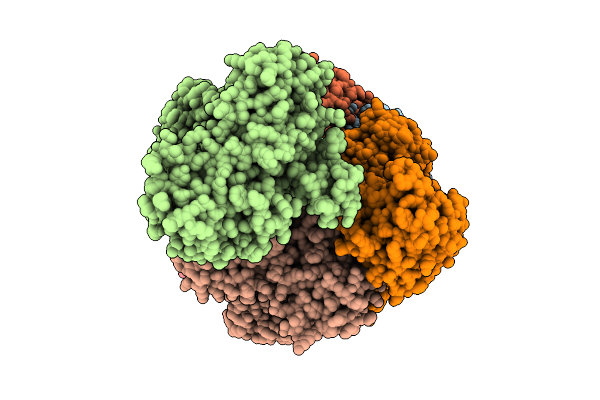 |
Cryoem Structure Of A Filamentous Soluble Pyridine Nucleotide Transhydrogenase
Organism: Azotobacter vinelandii
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: FLAVOPROTEIN Ligands: FAD |
 |
Organism: Azotobacter vinelandii
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: STRUCTURAL PROTEIN |
 |
Organism: Azotobacter vinelandii
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: METAL BINDING PROTEIN Ligands: HEM |
 |
Organism: Azotobacter vinelandii
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: OXIDOREDUCTASE Ligands: ICS, HCA, CLF, FE |
 |
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-12-11 Classification: HYDROLASE/INHIBITOR |
 |
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-12-11 Classification: HYDROLASE/INHIBITOR Ligands: Y33, GOL |
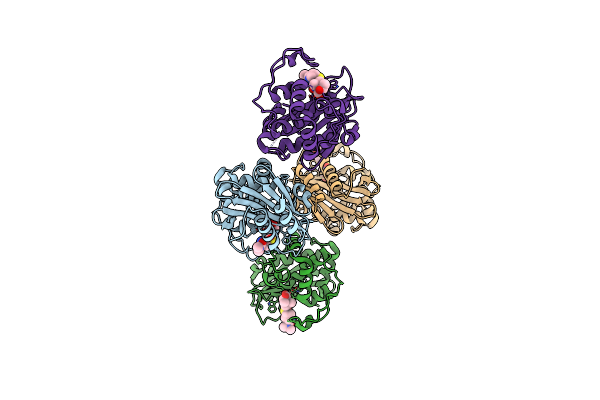 |
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2024-12-11 Classification: HYDROLASE/INHIBITOR Ligands: Y33 |
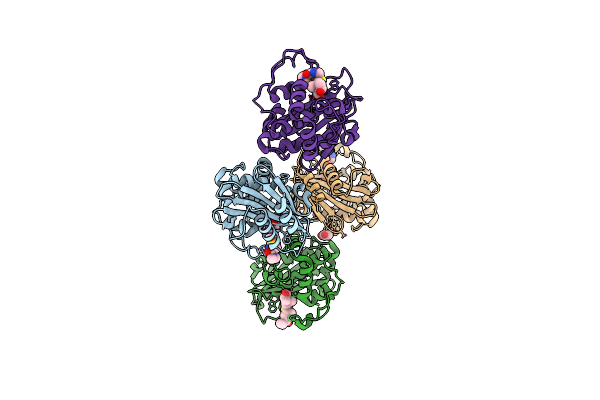 |
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2024-12-11 Classification: HYDROLASE/INHIBITOR Ligands: Y33, ACT |
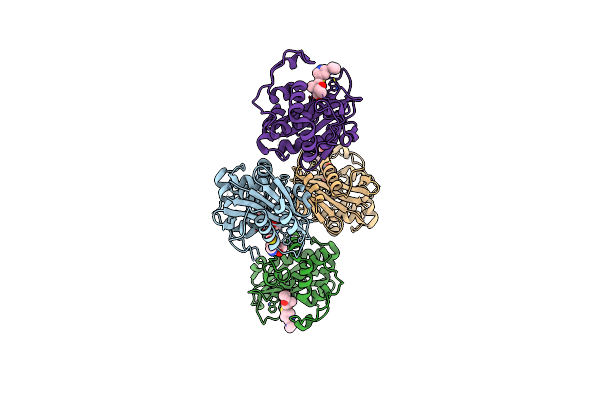 |
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2024-12-11 Classification: HYDROLASE/INHIBITOR Ligands: Y33, CO2 |
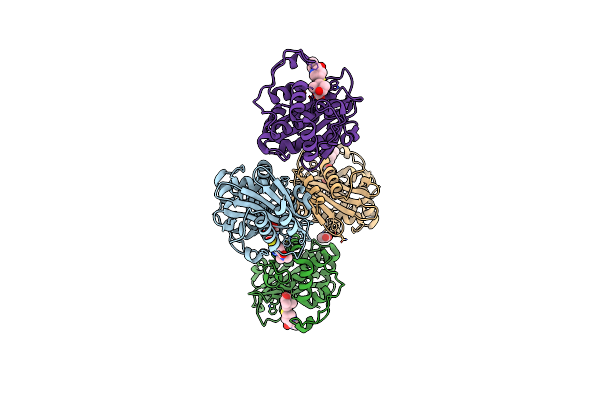 |
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2024-12-11 Classification: HYDROLASE/INHIBITOR Ligands: Y33, CO2, ACT |
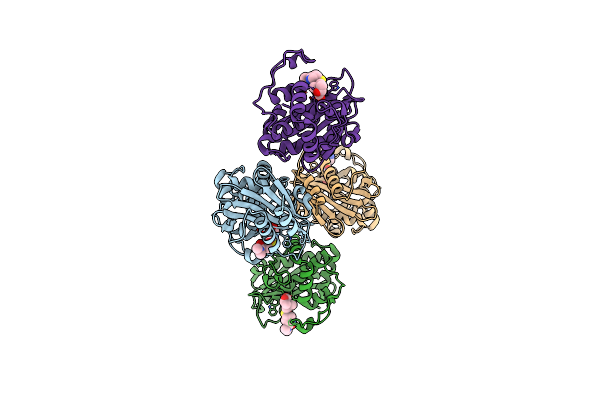 |
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2024-12-11 Classification: HYDROLASE/INHIBITOR Ligands: Y33 |
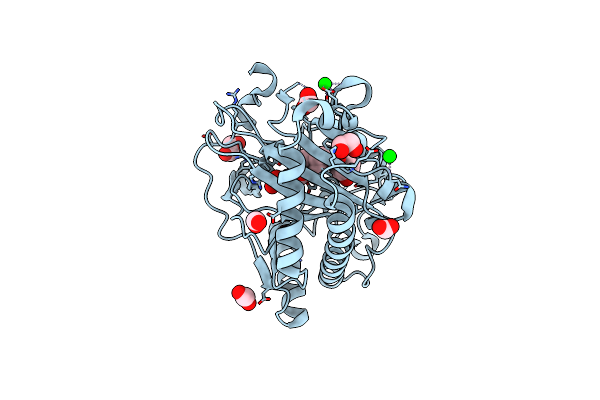 |
Structure Of Hyoscyamine 6-Beta Hydroxylase In Complex With Iron, 2-Oxoglutarate, And Hyoscyamine
Organism: Atropa belladonna
Method: X-RAY DIFFRACTION Resolution:1.53 Å Release Date: 2023-11-22 Classification: OXIDOREDUCTASE Ligands: SR, EDO, FMT, FE2, AKG, HYO |
 |
Structure Of Hyoscyamine 6-Beta Hydroxylase In Complex With Vanadyl, Succinate, And Hyoscyamine
Organism: Atropa belladonna
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2023-11-22 Classification: OXIDOREDUCTASE Ligands: EDO, HYO, SIN, FMT, V, O, SR |
 |
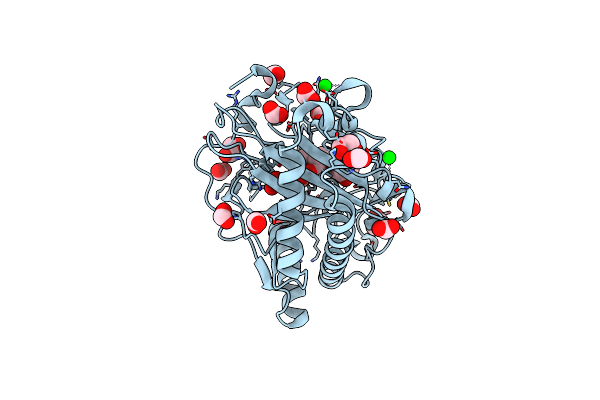
Structure Of Hyoscyamine 6-Beta Hydroxylase In Complex With Iron, Succinate, And 6-Oh-Hyoscyamine
Organism: Atropa belladonna
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2023-11-22 Classification: OXIDOREDUCTASE Ligands: FE2, EDO, FMT, OVR, SIN, SR |
 |
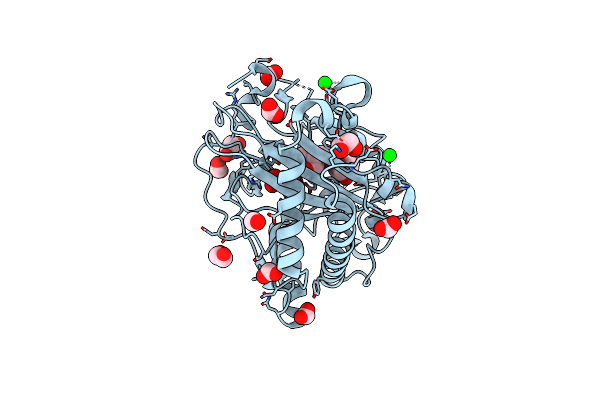
Structure Of Hyoscyamine 6-Beta Hydroxylase In Complex With Iron, 2-Oxoglutarate, And 6-Oh-Hyoscyamine
Organism: Atropa belladonna
Method: X-RAY DIFFRACTION Resolution:1.53 Å Release Date: 2023-11-22 Classification: OXIDOREDUCTASE Ligands: FE2, EDO, FMT, AKG, OVR, SR |
 |
Structure Of Hyoscyamine 6-Beta Hydroxylase In Complex With Vanadyl, Succinate, And 6-Oh-Hyoscyamine
Organism: Atropa belladonna
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2023-11-22 Classification: OXIDOREDUCTASE Ligands: EDO, SIN, FMT, OVR, V, O, SR |
 |
Structure Of Hyoscyamine 6-Beta Hydroxylase In Complex With Iron, Succinate, And Scopolamine
Organism: Atropa belladonna
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2023-11-22 Classification: OXIDOREDUCTASE Ligands: FE2, EDO, FMT, OW0, SIN, SR |

