Search Count: 36
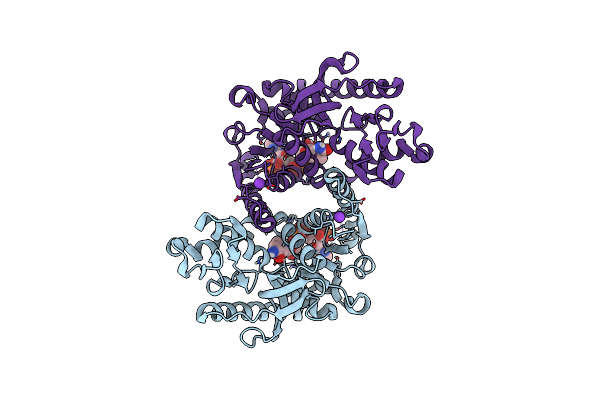 |
Crystal Structure Of Methanopyrus Kandleri Malate Dehydrogenase Mutant 4 At Room Temperature
Organism: Methanopyrus kandleri
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2025-03-19 Classification: OXIDOREDUCTASE Ligands: NDP, CL, K |
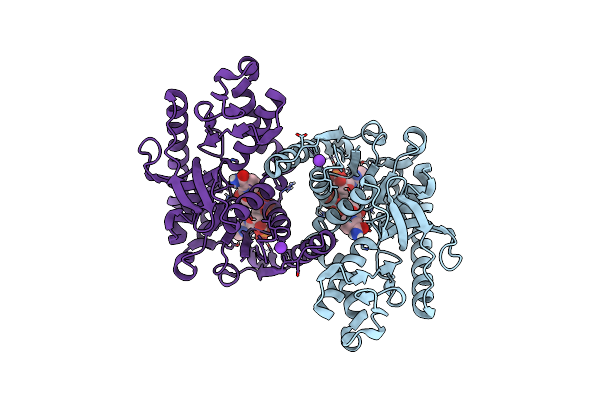 |
Organism: Methanopyrus kandleri
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2024-07-10 Classification: OXIDOREDUCTASE Ligands: NDP, CL, K |
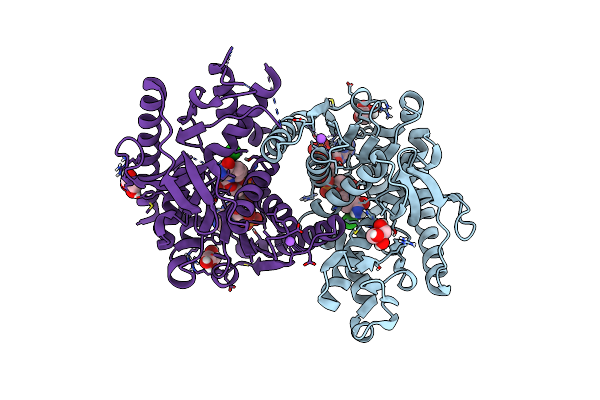 |
Organism: Methanopyrus kandleri
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2024-07-10 Classification: OXIDOREDUCTASE Ligands: NDP, GOL, CL, NA |
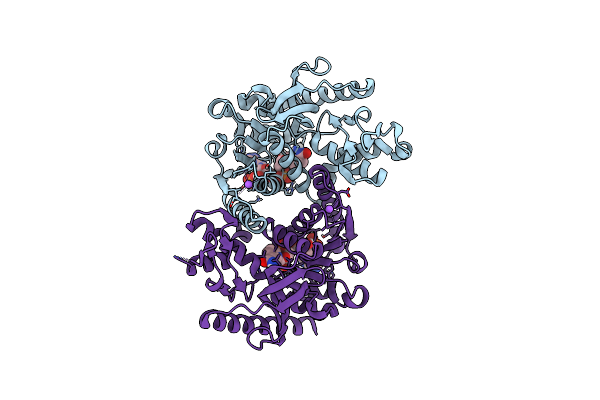 |
Organism: Methanopyrus kandleri
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2024-07-10 Classification: OXIDOREDUCTASE Ligands: NDP, CL, NA |
 |
Organism: Selenomonas ruminantium
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2023-11-01 Classification: OXIDOREDUCTASE Ligands: PO4, NO3, CL |
 |
Crystal Structure Of The Lactate Dehydrogenase Of Cyanobacterium Aponinum In Its Apo Form.
Organism: Cyanobacterium aponinum
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2022-08-03 Classification: OXIDOREDUCTASE Ligands: 7MT, TB, EDO |
 |
Crystal Structure Of The Lactate Dehydrogenase Of Cyanobacterium Aponinum In Complex With Oxamate, Nadh And Fbp.
Organism: Cyanobacterium aponinum
Method: X-RAY DIFFRACTION Resolution:2.62 Å Release Date: 2022-07-27 Classification: OXIDOREDUCTASE Ligands: OXM, FBP, NAI |
 |
Crystal Structure Of Malate Dehydrogenase From Haloarcula Marismortui With Potassium And Chloride Ions
Organism: Haloarcula marismortui
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2022-02-16 Classification: OXIDOREDUCTASE Ligands: CL, K |
 |
Crystal Structure Of Lactate Dehydrogenase From Selenomonas Ruminantium With Nadh.
Organism: Selenomonas ruminantium
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2021-02-10 Classification: OXIDOREDUCTASE Ligands: NAD, MLA, BO3, EDO, 1PE |
 |
Organism: Thermaerobacter marianensis
Method: X-RAY DIFFRACTION Resolution:2.12 Å Release Date: 2020-10-28 Classification: OXIDOREDUCTASE Ligands: GOL, PEG, 1PE, EDO, PGE |
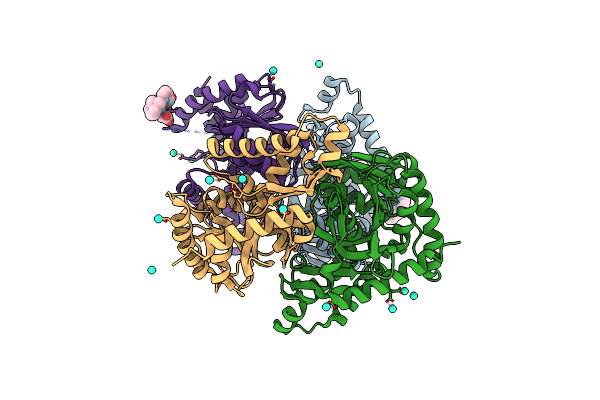 |
Crystal Structure Of Ignicoccus Islandicus Malate Dehydrogenase Co-Crystallized With 10 Mm Tb-Xo4
Organism: Ignicoccus islandicus dsm 13165
Method: X-RAY DIFFRACTION, SOLUTION SCATTERING Resolution:1.89 Å Release Date: 2019-07-17 Classification: OXIDOREDUCTASE Ligands: 7MT, TB, CL |
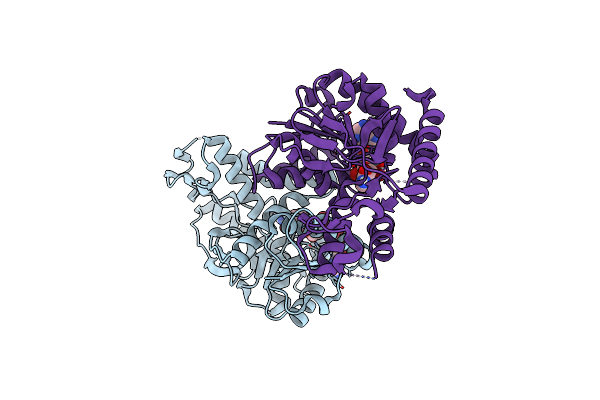 |
Crystal Dimeric Structure Of Petrotoga Mobilis Lactate Dehydrogenase With Nadh
Organism: Petrotoga mobilis (strain dsm 10674 / sj95)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2018-08-22 Classification: OXIDOREDUCTASE Ligands: NAD |
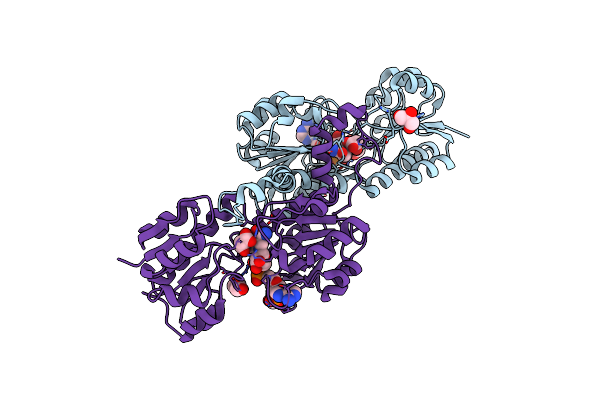 |
Crystal Structure Of Pyrococcus Yayanosii Glyoxylate Hydroxypyruvate Reductase In Complex With Nadp And Malonate (Re-Refinement Of 5Aow)
Organism: Pyrococcus yayanosii (strain ch1 / jcm 16557)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2018-01-17 Classification: OXIDOREDUCTASE Ligands: NAP, MLI, GOL |
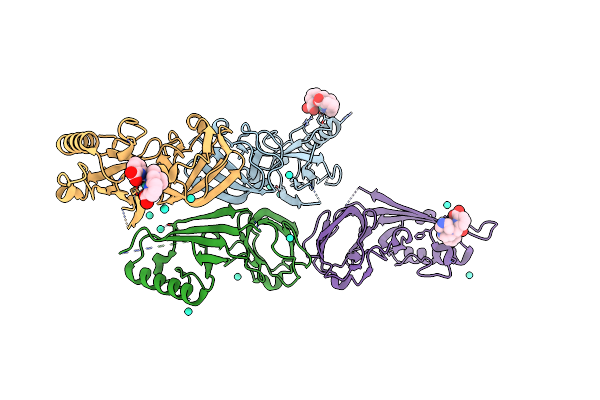 |
Organism: Escherichia phage t5
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-09-20 Classification: VIRAL PROTEIN Ligands: TB, 7MT, CL |
 |
Ternary Crystal Structure Of Pyrococcus Furiosus Glyoxylate Hydroxypyruvate Reductase In Presence Of Glyoxylate
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2016-03-02 Classification: OXIDOREDUCTASE Ligands: NAP, GLV, 1PE, EDO, ACY |
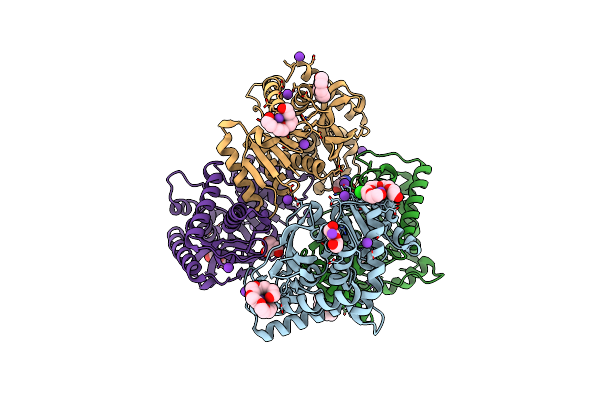 |
1.50 A Resolution Structure Of The Malate Dehydrogenase From Haloferax Volcanii
Organism: Haloferax volcanii
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2014-04-16 Classification: OXIDOREDUCTASE Ligands: K, CL, TRS, PGE, 1PE, P33, PG4 |
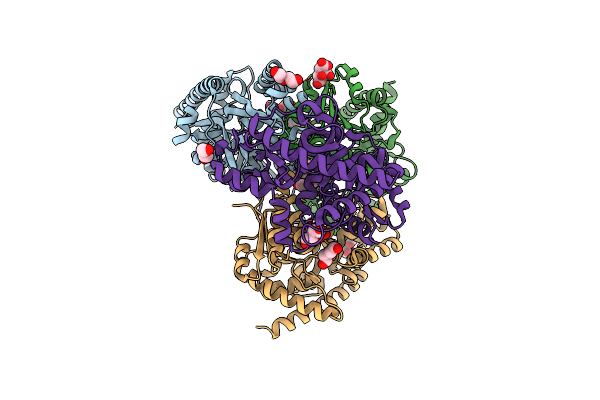 |
1.8 A Resolution Structure Of The Malate Dehydrogenase From Picrophilus Torridus In Its Apo Form
Organism: Picrophilus torridus
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2014-04-16 Classification: OXIDOREDUCTASE Ligands: PEG, CIT, PG6 |
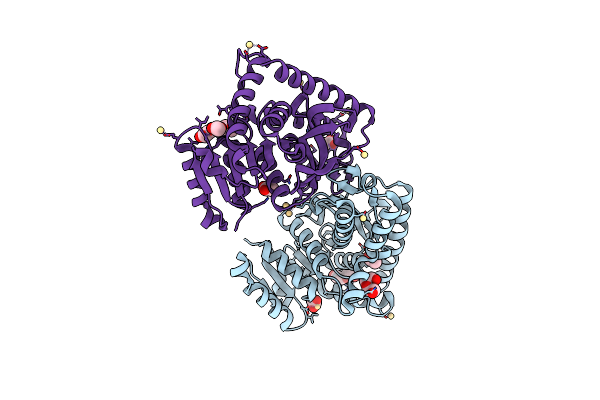 |
1.70 A Resolution Structure Of The Malate Dehydrogenase From Chloroflexus Aurantiacus
Organism: Chloroflexus sp. y-400-fl
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2014-02-05 Classification: OXIDOREDUCTASE Ligands: CD, CL, ACT, PEG |
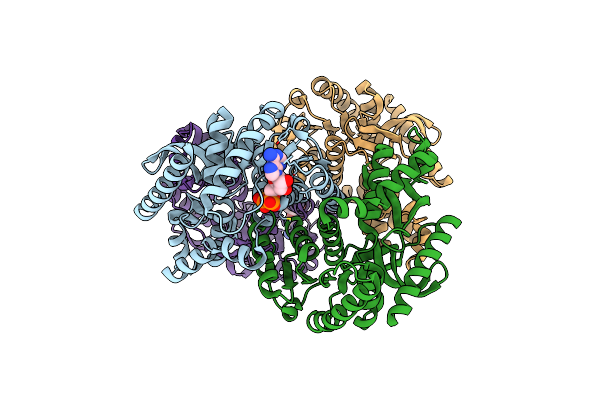 |
5-Mutant (R79W, R151A, E279A, E299A,E313A) Lactate-Dehydrogenase From Thermus Thermophillus
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2012-02-08 Classification: OXIDOREDUCTASE Ligands: ADP |
 |
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2011-12-28 Classification: OXIDOREDUCTASE |

