Search Count: 21
 |
Architecture Of A Pks-Nrps Hybrid Megaenzyme Involved In The Biosynthesis Of The Genotoxin Colibactin
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.98 Å Release Date: 2023-04-26 Classification: BIOSYNTHETIC PROTEIN |
 |
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2021-07-21 Classification: HYDROLASE Ligands: PO4 |
 |
Organism: Aequorea victoria
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2020-02-19 Classification: FLUORESCENT PROTEIN |
 |
Difference-Refined Structure Of Rsegfp2 10 Ns Following 400-Nm Laser Irradiation Of The Off-State Determined By Sfx
Organism: Aequorea victoria
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2020-02-19 Classification: FLUORESCENT PROTEIN |
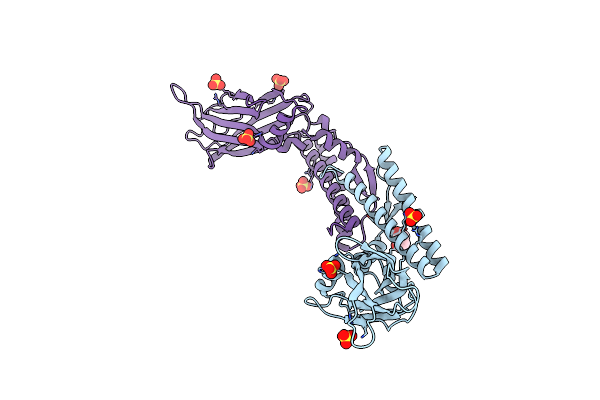 |
Cc2D1B Coordinates Esrct-Iii Activity During The Mitotic Reformation Of The Nuclear Envelope
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:2.46 Å Release Date: 2018-10-10 Classification: CYTOSOLIC PROTEIN Ligands: SO4, PEG |
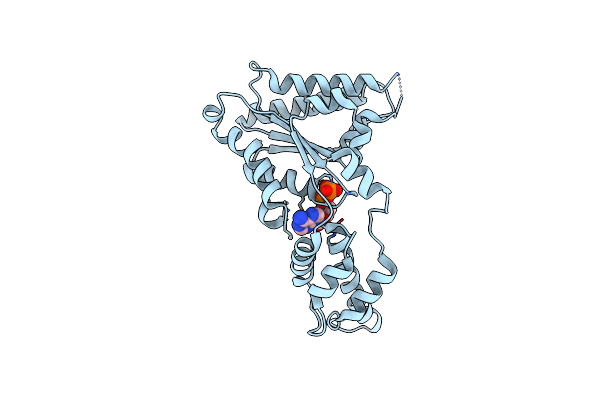 |
Organism: Metallosphaera sedula
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2015-11-25 Classification: HYDROLASE Ligands: ADP |
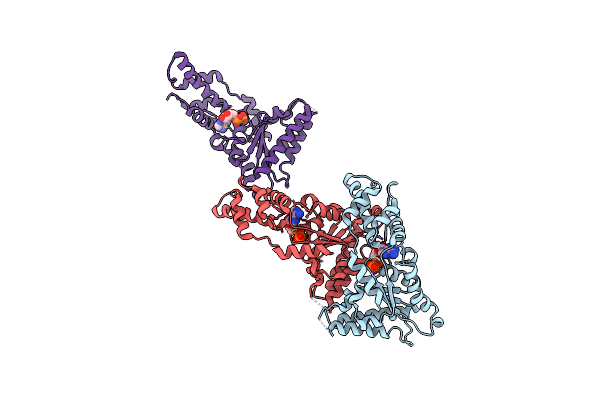 |
Organism: Metallosphaera sedula
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2015-11-25 Classification: HYDROLASE Ligands: ADP |
 |
Organism: Metallosphaera sedula
Method: X-RAY DIFFRACTION Resolution:3.60 Å Release Date: 2015-10-28 Classification: HYDROLASE |
 |
Organism: Lama glama
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2013-03-27 Classification: IMMUNE SYSTEM |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2011-08-24 Classification: HYDROLASE/PROTEIN TRANSPORT |
 |
Organism: Streptococcus pyogenes, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2011-04-13 Classification: CELL ADHESION |
 |
Organism: Streptococcus pyogenes, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:7.50 Å Release Date: 2011-04-13 Classification: CELL ADHESION |
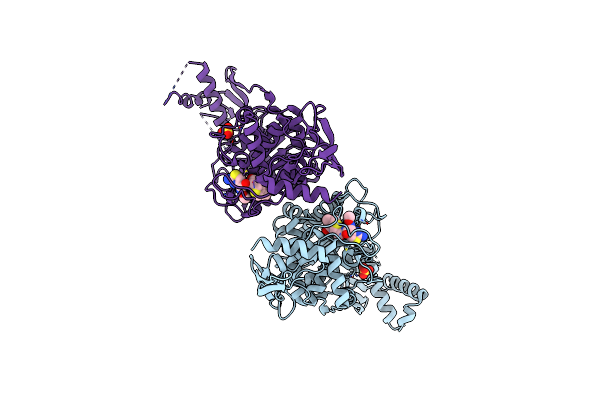 |
Active Site Restructuring Regulates Ligand Recognition In Class A Penicillin-Binding Proteins
Organism: Streptococcus pneumoniae
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2010-05-26 Classification: TRANSFERASE Ligands: CEF, SO4 |
 |
Structural Insights Into The Catalytic Mechanism And The Role Of Streptococcus Pneumoniae Pbp1B
Organism: Streptococcus pneumoniae
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2010-05-26 Classification: TRANSFERASE Ligands: SO4, S2D, CL |
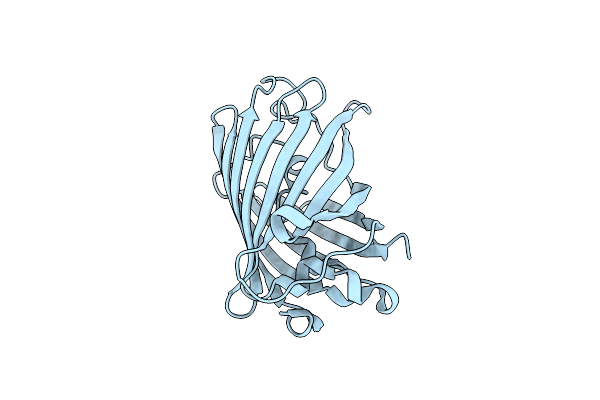 |
Organism: Aequorea victoria
Method: X-RAY DIFFRACTION Resolution:1.37 Å Release Date: 2009-09-29 Classification: FLUORESCENT PROTEIN |
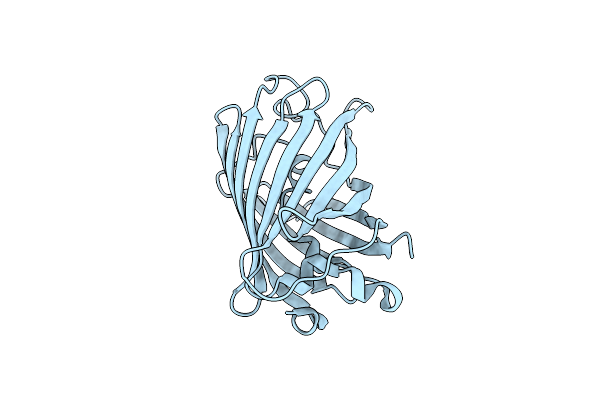 |
Organism: Aequorea victoria
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2009-09-29 Classification: FLUORESCENT PROTEIN |
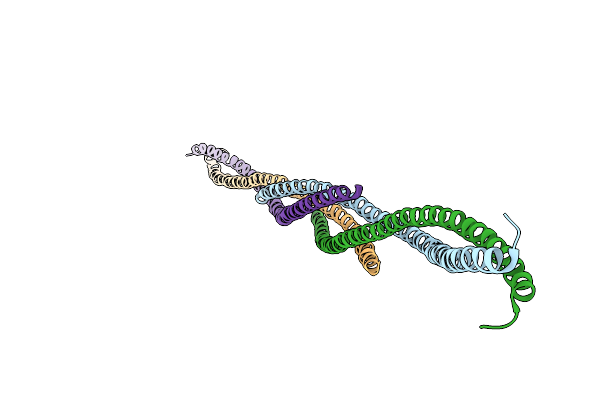 |
Organism: Streptococcus pyogenes serotype m1
Method: X-RAY DIFFRACTION Resolution:3.04 Å Release Date: 2008-03-18 Classification: SURFACE ACTIVE PROTEIN, TOXIN |
 |
Structural And Mechanistic Basis Of Penicillin Binding Protein Inhibition By Lactivicins
Organism: Streptococcus pneumoniae
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2007-08-14 Classification: DRUG-BINDING PROTEIN Ligands: SO4, CL, EDO, PL7 |
 |
Structural And Mechanistic Basis Of Penicillin Binding Protein Inhibition By Lactivicins
Organism: Streptococcus pneumoniae
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2007-08-14 Classification: DRUG-BINDING PROTEIN Ligands: L4C, SO4, CL |
 |
Active Site Restructuring Regulates Ligand Recognition In Class A Penicillin-Binding Proteins
Organism: Streptococcus pneumoniae
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2007-04-03 Classification: TRANSFERASE Ligands: SO4, CL, EDO |

