Search Count: 7
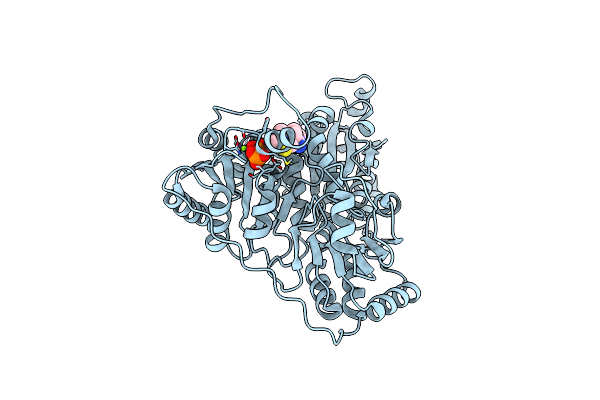 |
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:2.53 Å Release Date: 2024-12-11 Classification: BIOSYNTHETIC PROTEIN Ligands: TPP, MG |
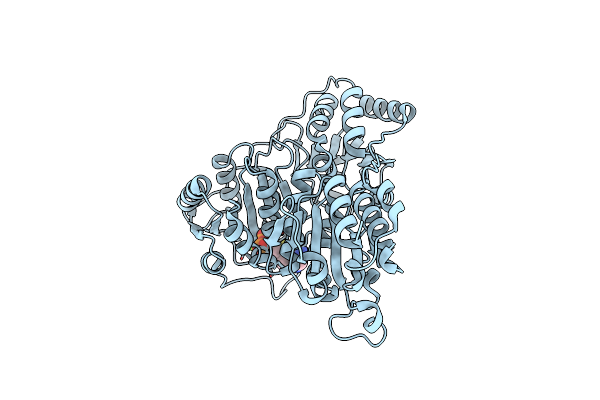 |
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2024-12-11 Classification: BIOSYNTHETIC PROTEIN Ligands: TPP, MG |
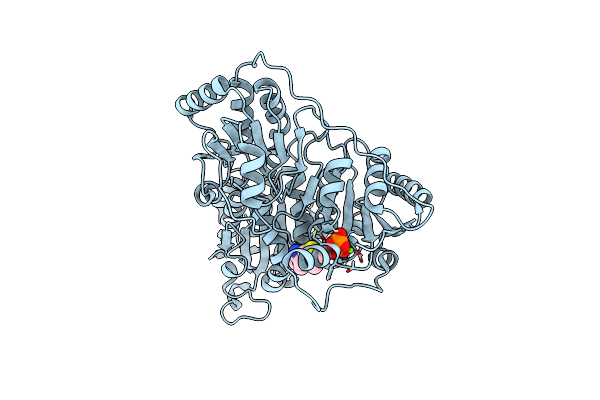 |
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2024-12-11 Classification: BIOSYNTHETIC PROTEIN Ligands: TPP, MG |
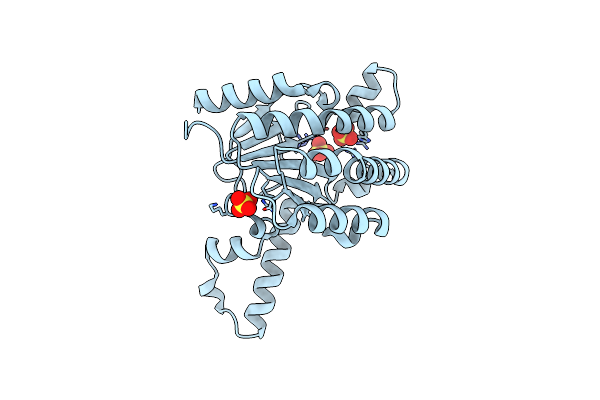 |
Crystal Structure Of Lavandulyl Diphosphate Synthase From Lavandula X Intermedia In Apo Form
Organism: Lavandula lanata
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2016-03-02 Classification: TRANSFERASE Ligands: SO4 |
 |
Crystal Structure Of Lavandulyl Diphosphate Synthase From Lavandula X Intermedia In Complex With S-Thiolo-Isopentenyldiphosphate
Organism: Lavandula lanata
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2016-03-02 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: MG, DST |
 |
Crystal Structure Of Lavandulyl Diphosphate Synthase From Lavandula X Intermedia In Complex With Dimethylallyl Diphosphate
Organism: Lavandula lanata
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2016-03-02 Classification: TRANSFERASE Ligands: MG, PIS, 61G, SO4, ISY |
 |
Crystal Structure Of Endo-Beta-1,4-Xylanase From The Alkaliphilic Bacillus Sp. Sn5
Organism: Bacillus sp
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2014-02-12 Classification: HYDROLASE Ligands: SO4 |

