Search Count: 1,387
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: PROTEIN FIBRIL |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: PROTEIN FIBRIL |
 |
Organism: Euglena gracilis
Method: ELECTRON MICROSCOPY Release Date: 2025-10-08 Classification: PHOTOSYNTHESIS Ligands: CL0, CLA, PQN, SF4, BCR, LHG, UNL, DGD, DD6 |
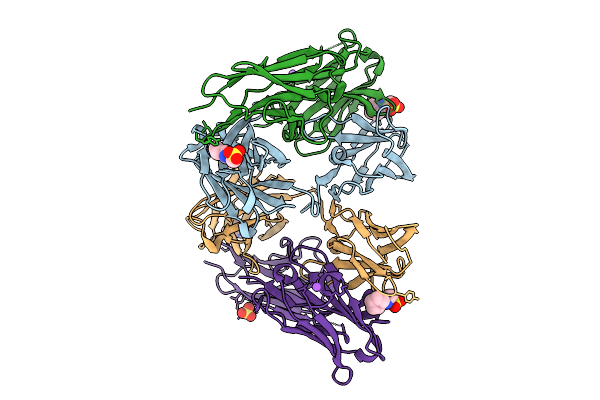 |
Crystal Structure Of A High Affinity Vl-Vh Tetrabody For The Erythropoietin Receptor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: SIGNALING PROTEIN Ligands: NHE, CL, SO4, NA |
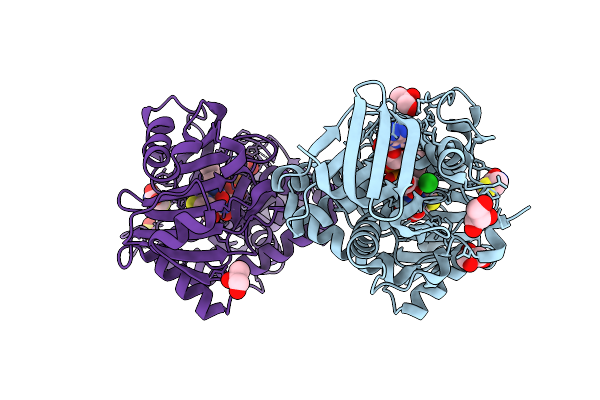 |
Monomeric Sarcosine Oxidase From Bacillus Sp. (Soxb) Complexed With L-Thioproline
Organism: Bacillus sp. b-0618
Method: X-RAY DIFFRACTION Release Date: 2025-09-24 Classification: OXIDOREDUCTASE Ligands: FAD, PRS, CL, GOL, SO4 |
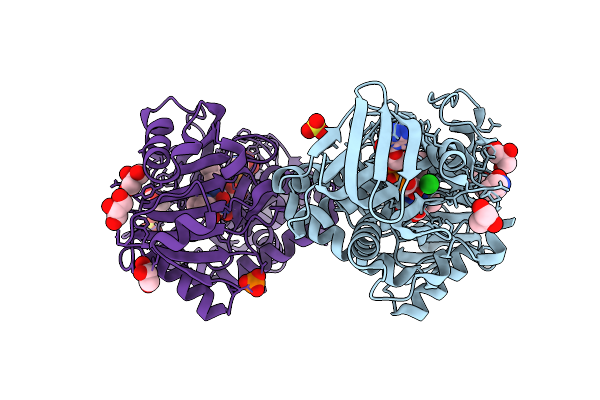 |
Monomeric Sarcosine Oxidase From Bacillus Sp. (Soxb) Complexed With L-Proline
Organism: Bacillus sp. b-0618
Method: X-RAY DIFFRACTION Release Date: 2025-09-24 Classification: OXIDOREDUCTASE Ligands: FAD, PRO, GOL, CL, SO4, PO4 |
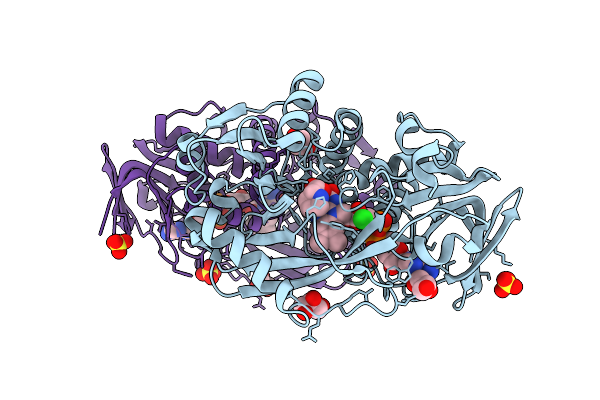 |
Monomeric Sarcosine Oxidase From Bacillus Sp. (Soxb) Complexed With D-Proline
Organism: Bacillus sp. b-0618
Method: X-RAY DIFFRACTION Release Date: 2025-09-24 Classification: OXIDOREDUCTASE Ligands: FAD, DPR, CL, GOL, SO4 |
 |
Organism: Bacillus sp. b-0618
Method: X-RAY DIFFRACTION Release Date: 2025-09-24 Classification: OXIDOREDUCTASE Ligands: FAD, GOL, CL, NA |
 |
Organism: Bacillus sp. b-0618
Method: X-RAY DIFFRACTION Release Date: 2025-09-24 Classification: OXIDOREDUCTASE Ligands: FAD, GOL, CL |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-08-27 Classification: MEMBRANE PROTEIN Ligands: PTY |
 |
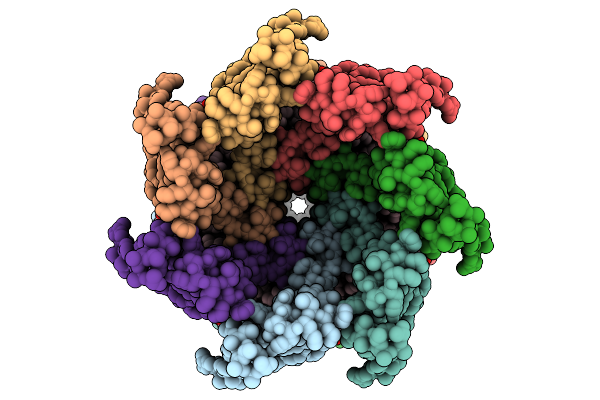
Cryo-Em Structure Of Mechanosensitive Channel Ynai In Dopc Nanodiscs Treated With Beta-Cyclodextrin
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-08-27 Classification: MEMBRANE PROTEIN Ligands: PTY |
 |
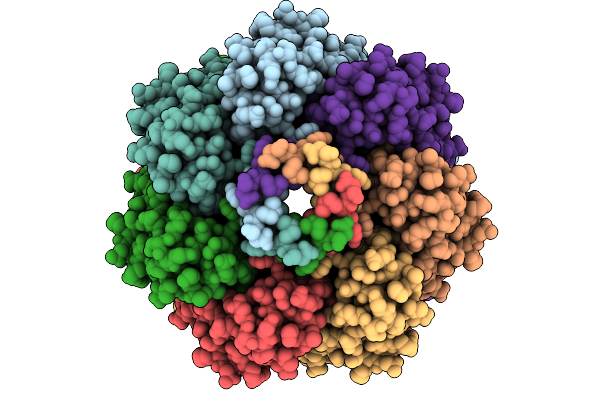
Cryo-Em Structure Of Mechanosensitive Channel Ynai In Dopc Nanodiscs Treated With Lysophosphatidylcholine
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-08-27 Classification: MEMBRANE PROTEIN Ligands: PTY |
 |
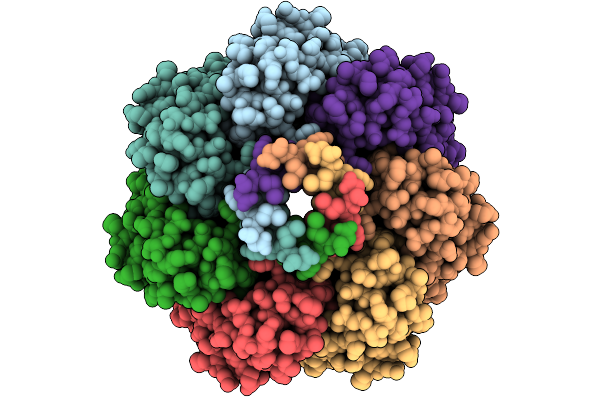
Cryo-Em Structure Of Mechanosensitive Channel Ynai A155V Mutant In Conformation 1
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-08-27 Classification: MEMBRANE PROTEIN Ligands: PTY |
 |
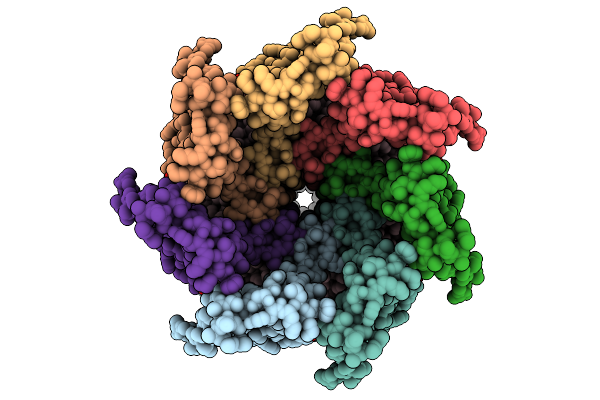
Cryo-Em Structure Of Mechanosensitive Channel Ynai A155V Mutant In Conformation 2
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-08-27 Classification: MEMBRANE PROTEIN Ligands: PTY |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-08-27 Classification: MEMBRANE PROTEIN Ligands: PTY |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-08-27 Classification: MEMBRANE PROTEIN |
 |
Crystal Structure Of Hla-C*12:02 In Complex With Kaynvtqaf (Kf9), A 9-Mer Epitope From Sars-Cov-2 Nucleocapsid (N266-274)
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: IMMUNE SYSTEM |
 |
Crystal Structure Of Gv37-Tcr In Complex With Hla-C*12:02 With Kaynvtqaf (Kf9), A 9-Mer Epitope From Sars-Cov-2 Nucleocapsid (N266-274)
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: IMMUNE SYSTEM Ligands: EDO, SO4 |
 |
Crystal Structure Of Human 1122A11 Fab In Complex With Influenza Virus Neuraminidase From A/Singapore/Infimh-16-0019/2016 (H3N2)
Organism: Influenza a virus, Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: VIRAL PROTEIN, HYDROLASE/IMMUNE SYSTEM Ligands: CA, NAG |
 |
Crystal Structure Of Influenza Virus N2 Neuraminidase From A/Singapore/Infimh-16-0019/2016 (H3N2)
Organism: Influenza a virus
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: VIRAL PROTEIN/HYDROLASE Ligands: CA, SO4, NAG |

