Search Count: 1,938
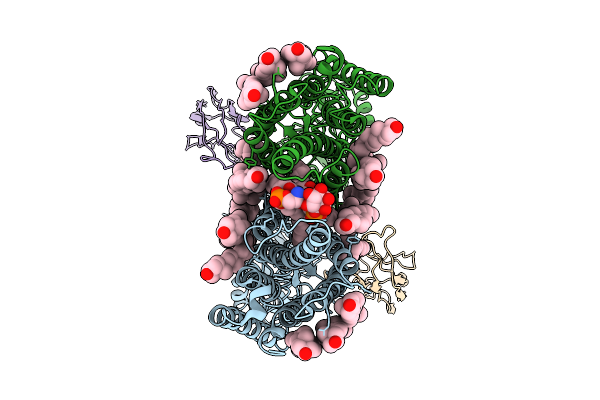 |
Organism: Homo sapiens, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: MEMBRANE PROTEIN Ligands: CLR, PEE, PIE |
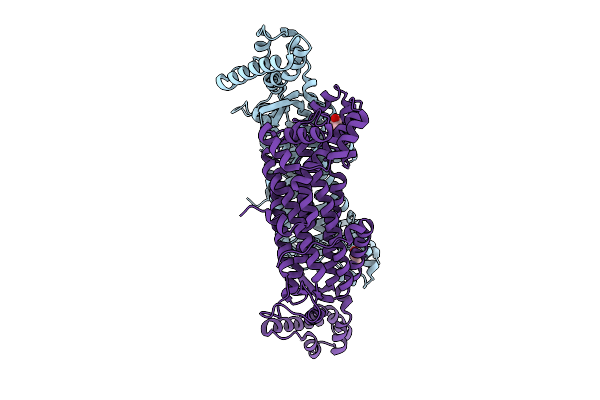 |
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: LYASE Ligands: GOL |
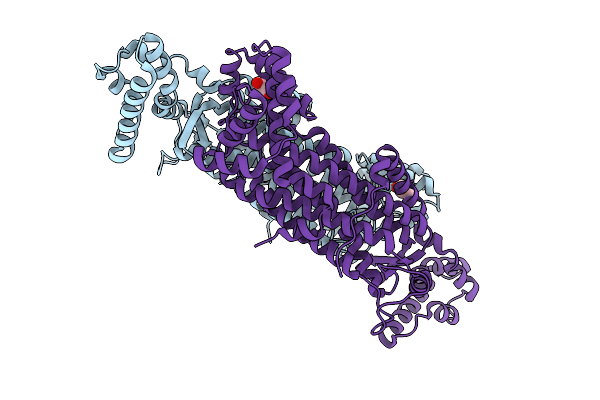 |
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: LYASE Ligands: GOL |
 |
Organism: Sus scrofa domesticus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: HYDROLASE Ligands: NAG |
 |
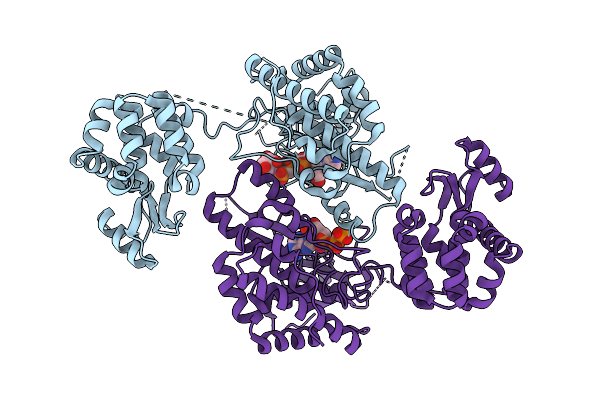
The Cryo-Em Structure Of Porcine Serum Mgam Bound With Acarviosyl-Maltotriose.
Organism: Sus scrofa domesticus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: HYDROLASE Ligands: NAG |
 |
Crystal Structure Of Phosphatidyl Inositol 4-Kinase Ii Beta In Complex With Hh5129
Organism: Homo sapiens, Enterobacteria phage t4
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: TRANSFERASE Ligands: A1IVA |
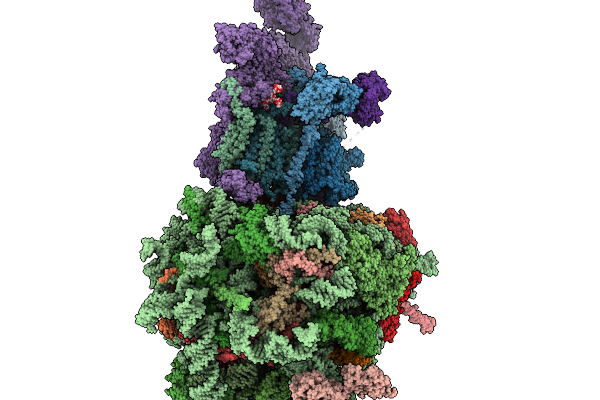 |
Structure Of A Grp94 Folding Intermediate Engaged With A Ccdc134- And Fkbp11-Bound Secretory Translocon
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: RIBOSOME Ligands: ELU, MG, ZN |
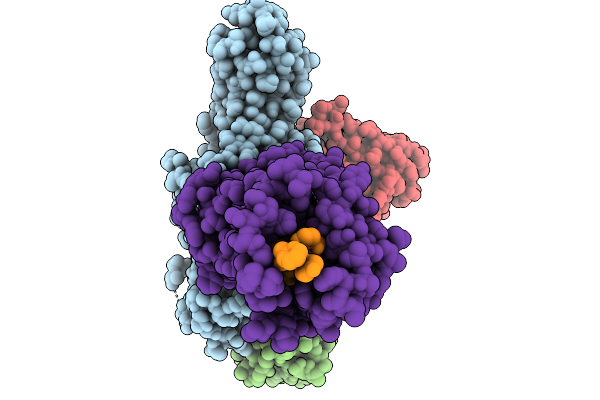 |
Cryo-Em Structure Of The G Protein-Coupled Receptor 1 (Gpr1) Bound To Chemerin And Beta-Arrestin 1 (Conformation 1)
Organism: Homo sapiens, Escherichia phage ecszw-2
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: MEMBRANE PROTEIN/IMMUNE SYSTEM |
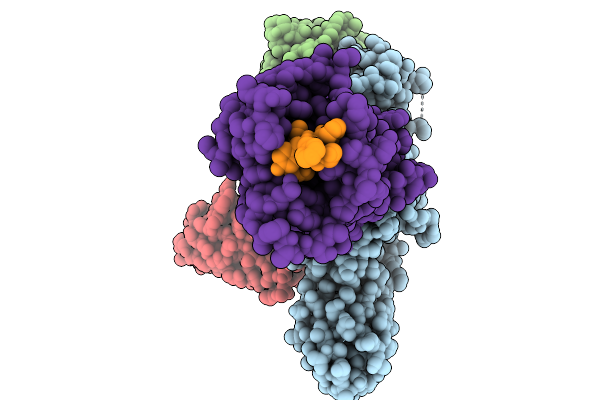 |
Cryo-Em Structure Of The G Protein-Coupled Receptor 1 (Gpr1) Bound To Chemerin And Beta-Arrestin 1 (Conformation 2)
Organism: Homo sapiens, Escherichia phage ecszw-2
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: MEMBRANE PROTEIN/IMMUNE SYSTEM |
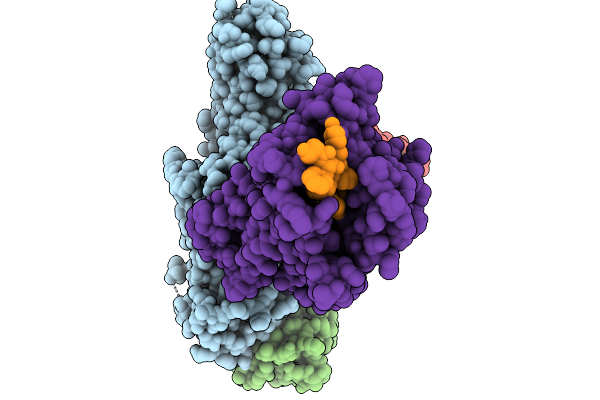 |
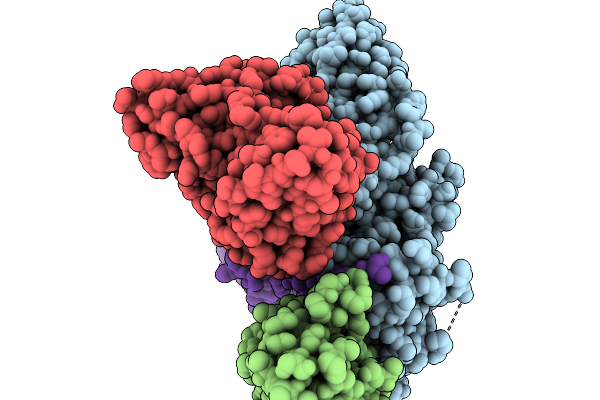
Cryo-Em Structure Of The G Protein-Coupled Receptor 1 (Gpr1) Bound To Chemerin And Beta-Arrestin 1 (Conformation 3)
Organism: Homo sapiens, Escherichia phage ecszw-2
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: MEMBRANE PROTEIN/IMMUNE SYSTEM |
 |
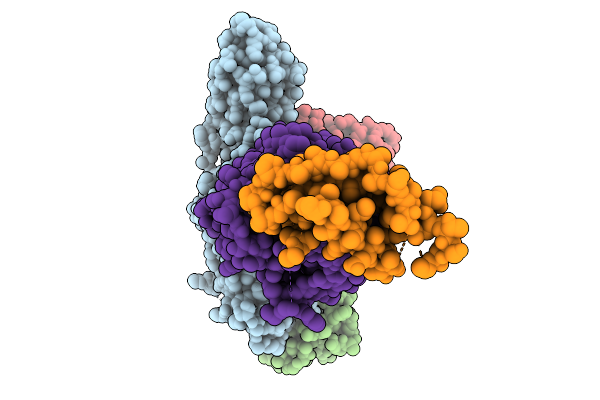
Cryo-Em Structure Of The G Protein-Coupled Receptor 1 (Gpr1) Bound To Chemerin And Beta-Arrestin 1 (Conformation 4)
Organism: Homo sapiens, Escherichia phage ecszw-2
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: MEMBRANE PROTEIN/IMMUNE SYSTEM |
 |
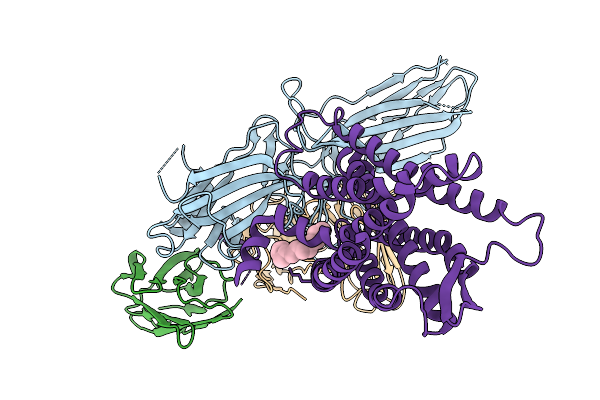
Composite Map Of The G Protein-Coupled Receptor 1 (Gpr1) Bound To Chemerin And Beta-Arrestin 2
Organism: Homo sapiens, Escherichia phage ecszw-2
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: MEMBRANE PROTEIN/IMMUNE SYSTEM Ligands: Y01 |
 |
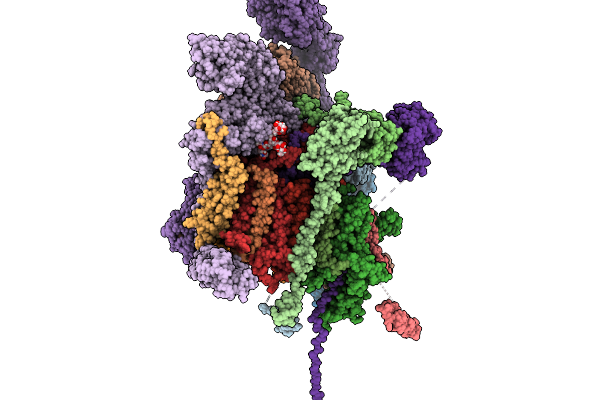
Cryo-Em Structure Of The G Protein-Coupled Receptor 1 (Gpr1) Bound To Beta-Arrestin 1 In Ligand-Free State
Organism: Homo sapiens, Escherichia phage ecszw-2
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: MEMBRANE PROTEIN/IMMUNE SYSTEM Ligands: PAM |
 |
Structure Of A Grp94 Folding Intermediate Engaged With A Ccdc134- And Fkbp11-Bound Secretory Translocon
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: RIBOSOME Ligands: ELU |
 |
Organism: Kribbella flavida
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: OXIDOREDUCTASE Ligands: NAP, SO4 |
 |
Imine Reductase Ir91 From Kribbella Flavida With Nadp+ And 5-Methoxy-2-Tetralone
Organism: Kribbella flavida
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: OXIDOREDUCTASE Ligands: SO4, NAP, A1JDH, EDO |
 |
Imine Reductase Ir91 From Kribbella Flavida With Nadp+ And 5-Methoxy-(S)-2-(N-Methylamino)Tetralin
Organism: Kribbella flavida
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: OXIDOREDUCTASE Ligands: SO4, NAP, A1JDG |
 |
Crystal Structure Of Bip Atpase Domain In Complex With Cdnf C-Terminal Domain At 1.65 Angstroms Resolution
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: CHAPERONE Ligands: MG, CL, PO4, GOL |
 |
Cryo-Em Structure Of Tmprss2 In Complex With Fab Fragments Of 752 Mab And 2228 Mab
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-10-15 Classification: MEMBRANE PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-15 Classification: IMMUNE SYSTEM Ligands: NAG |

