Search Count: 1,269
 |
Organism: Mus musculus, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: RNA BINDING PROTEIN/RNA Ligands: PEG |
 |
Staphylococcus Aureus Moaa C-Terminal Tail Peptide In Complex With G340A-Moaa
Organism: Staphylococcus aureus
Method: SOLUTION NMR Release Date: 2025-11-26 Classification: BIOSYNTHETIC PROTEIN |
 |
Cryo-Em Structure Of Human Kappa Opioid Receptor - G Protein Signaling Complex Bound With Nalfurafine.
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: MEMBRANE PROTEIN Ligands: IVB |
 |
Cryo-Em Structure Of Human Kappa Opioid Receptor -G Protein Signaling Complex Bound With U-50488H
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: MEMBRANE PROTEIN Ligands: A1LWX |
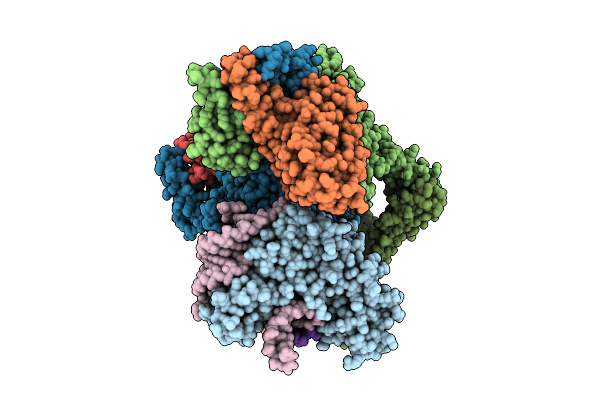 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: RNA BINDING PROTEIN Ligands: ADP, ZN, MG |
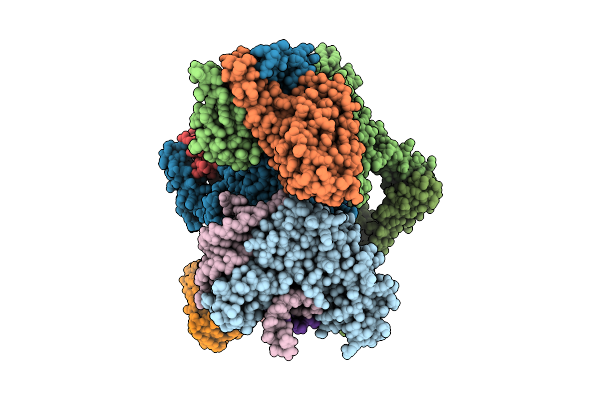 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: RNA BINDING PROTEIN Ligands: ADP, ZN, MG |
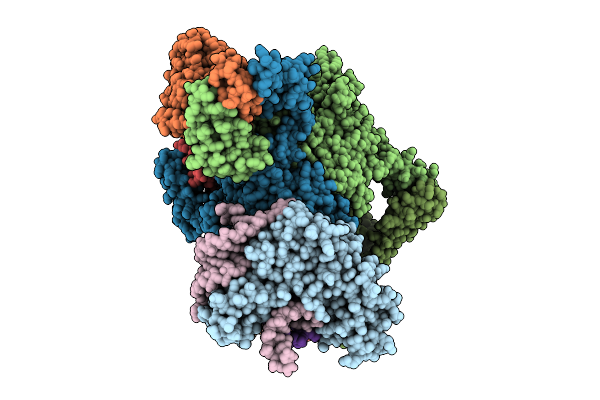 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: RNA BINDING PROTEIN Ligands: ADP, ZN, MG |
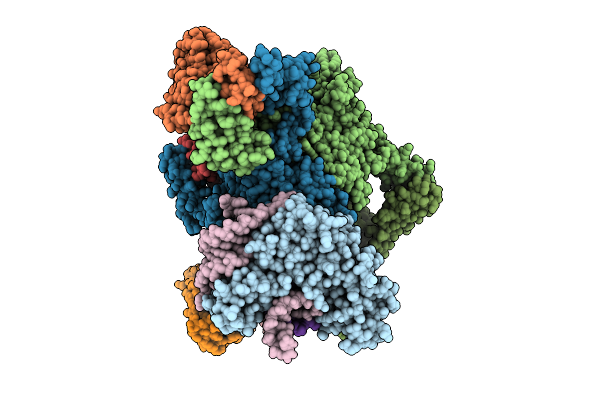 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: RNA BINDING PROTEIN Ligands: ADP, ZN, MG |
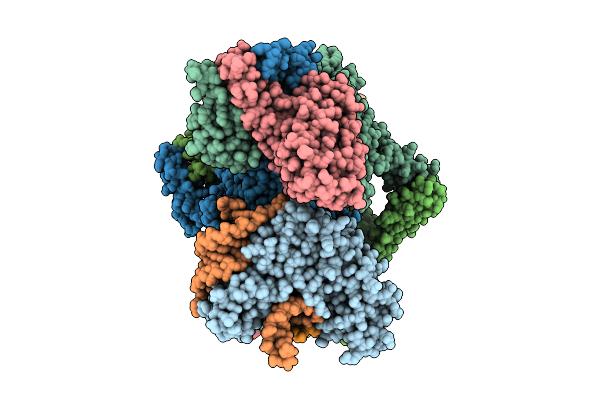 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: RNA BINDING PROTEIN Ligands: ADP, ZN, MG |
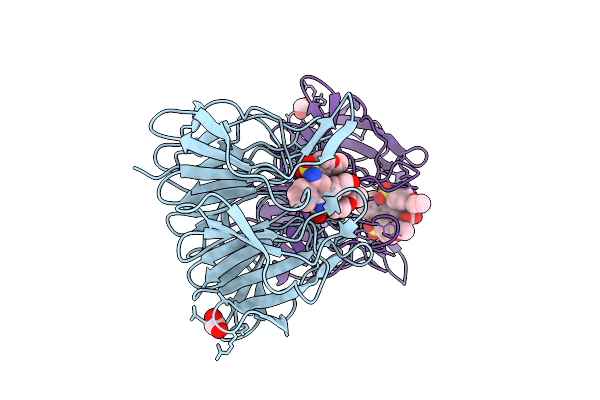 |
Crystal Structure Of Keap1 In Complex With A Small Molecule Inhibitor, Kmn003
Organism: Mus musculus
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: PEPTIDE BINDING PROTEIN/INHIBITOR Ligands: A1L98, FMT |
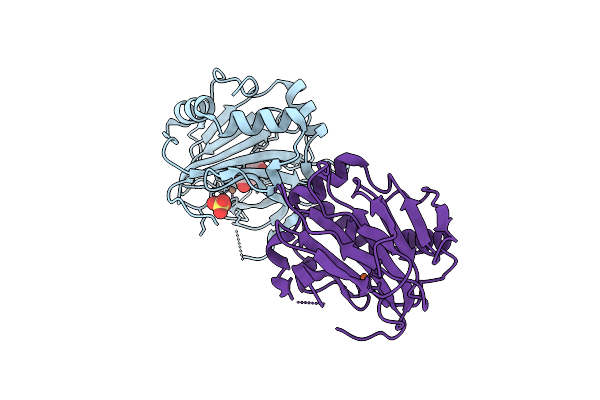 |
X-Ray Crystal Structure Of Streptomyces Cacaoi Pold With Iron And Succinate Bound
Organism: Streptomyces cacaoi
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: OXIDOREDUCTASE Ligands: FE, SO4, SIN |
 |
Organism: Thermus thermophilus hb8
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: MOTOR PROTEIN Ligands: MG, ADP, ATP |
 |
Organism: Thermus thermophilus hb8
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: MOTOR PROTEIN |
 |
Organism: Thermus thermophilus hb8
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: MOTOR PROTEIN |
 |
Organism: Thermus thermophilus hb8
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: MOTOR PROTEIN |
 |
Cryo-Em Structure Of The Vo Domain Of V/A-Atpase Under Pmf Condition, State3A
Organism: Thermus thermophilus hb8
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: MOTOR PROTEIN |
 |
Cryo-Em Structure Of The Vo Domain Of V/A-Atpase Under Pmf Condition, State3B
Organism: Thermus thermophilus hb8
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: MOTOR PROTEIN |
 |
Cryo-Em Structure Of The Vo Domain Of V/A-Atpase Under Pmf Condition, State3C
Organism: Thermus thermophilus hb8
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: MOTOR PROTEIN |
 |
Cryo-Em Structure Of The Vo Domain Of V/A-Atpase Under Pmf Condition, State3D
Organism: Thermus thermophilus hb8
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: MOTOR PROTEIN |
 |
Cryo-Em Structure Of The V1 Domain Of V/A-Atpase In Liposomes Under No Pmf Condition, State1W
Organism: Thermus thermophilus hb8
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: MOTOR PROTEIN Ligands: MG, ADP, PO4 |

