Search Count: 832
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2026-01-28 Classification: RNA BINDING PROTEIN Ligands: A1EY1, ECC |
 |
Organism: Gloeobacter violaceus pcc 7421
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: PHOTOSYNTHESIS Ligands: CYC |
 |
Organism: Thermus phage philo
Method: ELECTRON MICROSCOPY Resolution:3.81 Å Release Date: 2025-12-10 Classification: RNA BINDING PROTEIN/RNA |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.21 Å Release Date: 2025-12-10 Classification: IMMUNE SYSTEM |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:2.91 Å Release Date: 2025-12-10 Classification: IMMUNE SYSTEM |
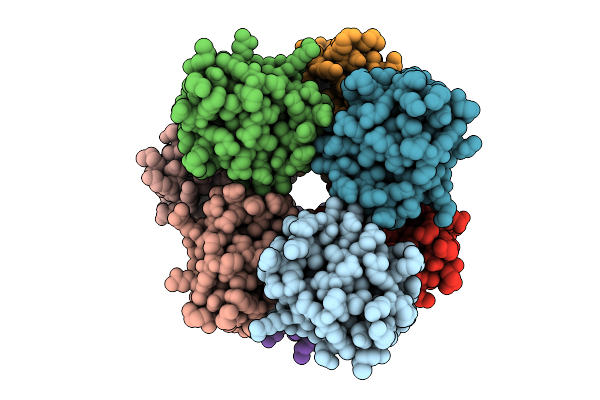 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:2.66 Å Release Date: 2025-12-10 Classification: IMMUNE SYSTEM |
 |
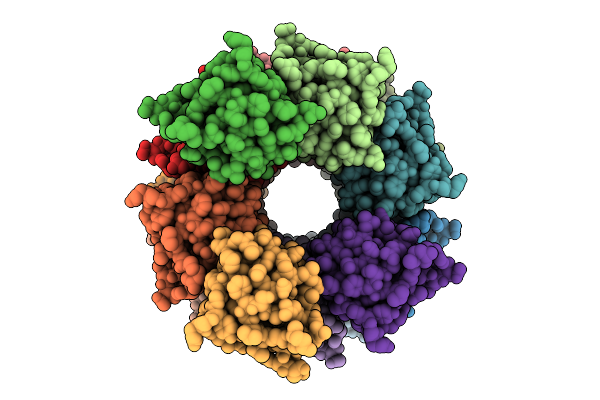
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:2.66 Å Release Date: 2025-12-10 Classification: IMMUNE SYSTEM |
 |
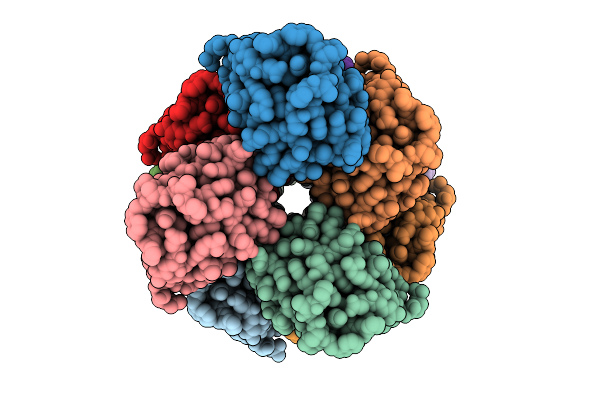
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.14 Å Release Date: 2025-12-10 Classification: IMMUNE SYSTEM |
 |
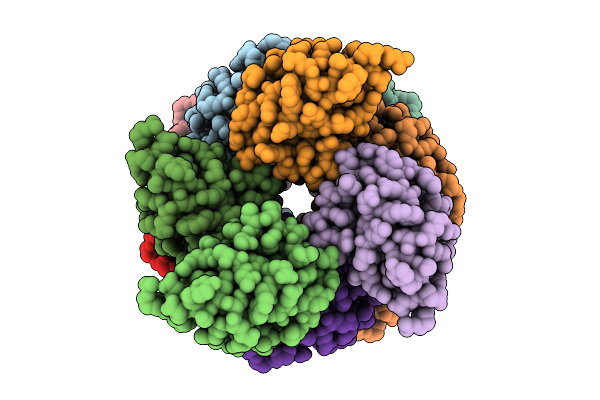
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:2.67 Å Release Date: 2025-12-10 Classification: IMMUNE SYSTEM |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:2.45 Å Release Date: 2025-12-10 Classification: IMMUNE SYSTEM |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:2.86 Å Release Date: 2025-12-10 Classification: IMMUNE SYSTEM |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:4.41 Å Release Date: 2025-12-10 Classification: IMMUNE SYSTEM |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:4.02 Å Release Date: 2025-12-10 Classification: IMMUNE SYSTEM |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:4.00 Å Release Date: 2025-12-10 Classification: IMMUNE SYSTEM |
 |
Organism: Gloeobacter violaceus pcc 7421
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: PHOTOSYNTHESIS Ligands: CYC |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-11-26 Classification: TRANSFERASE Ligands: SO4, A1EF6, EPE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.92 Å Release Date: 2025-11-26 Classification: TRANSFERASE Ligands: 6Y3, SO4, GOL, EPE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2025-11-26 Classification: TRANSFERASE Ligands: EPE, A1EF7 |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: TRANSFERASE Ligands: ATP, MG, BCT, 1VU |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: TRANSFERASE Ligands: ATP, MG, BCT, 1VU |

