Search Count: 65
 |
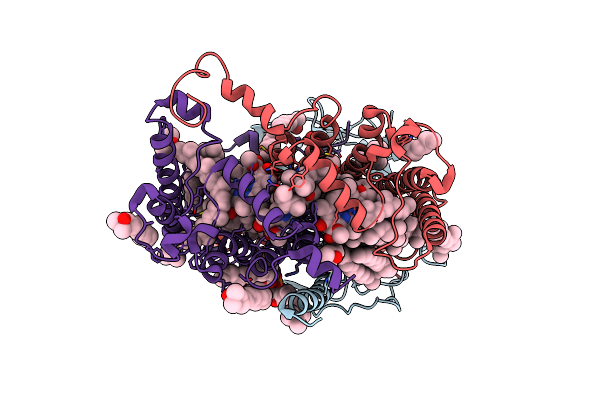
Structure Of The Human Dopamine Transporter In Complex With Beta-Cft, Mrs7292 And Divalent Zinc
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-08-07 Classification: MEMBRANE PROTEIN Ligands: 42L, A1AAF, NAG, Y01, D10, LNK, HP6, ZN, NA, D12, MYS, OCT |
 |
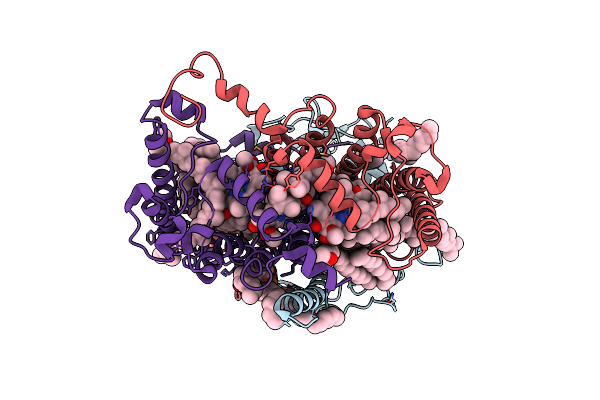
Structure Of Photosynthetic Reaction Center From Rhodobacter Sphaeroides Strain Rv By Fixed-Target Serial Synchrotron Crystallography (Room Temperature, 26Kev)
Organism: Cereibacter sphaeroides 2.4.1
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2023-03-29 Classification: PHOTOSYNTHESIS Ligands: LDA, BCL, BPH, D12, MYS, OLC, NKP, U10, FE, SPN, EDO, PO4 |
 |
F(M197)H Mutant Structure Of Photosynthetic Reaction Center From Rhodobacter Sphaeroides Strain Rv By Fixed-Target Serial Synchrotron Crystallography (Room Temperature, 12Kev)
Organism: Rhodobacter sphaeroides
Method: X-RAY DIFFRACTION Resolution:2.22 Å Release Date: 2022-07-13 Classification: PHOTOSYNTHESIS Ligands: MYS, LDA, OLC, BCL, BPH, NKP, U10, SPN, FE, EDO |
 |
F(M197)H Mutant Structure Of Photosynthetic Reaction Center From Rhodobacter Sphaeroides Strain Rv By Fixed-Target Serial Synchrotron Crystallography (Room Temperature, 26Kev)
Organism: Rhodobacter sphaeroides (strain atcc 17023 / dsm 158 / jcm 6121 / nbrc 12203 / ncimb 8253 / ath 2.4.1.)
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2022-04-27 Classification: PHOTOSYNTHESIS Ligands: LDA, OLC, MYS, BCL, BPH, NKP, U10, FE, SPN, EDO, PO4 |
 |
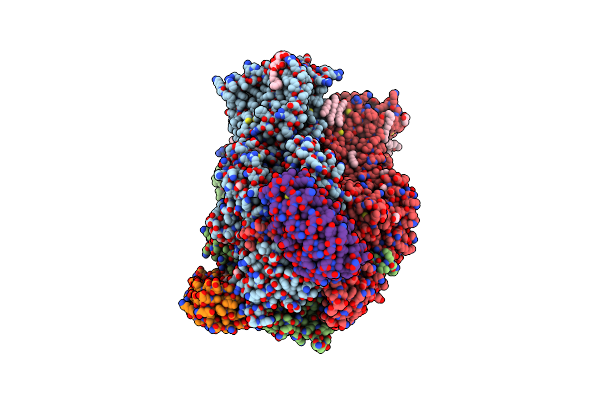
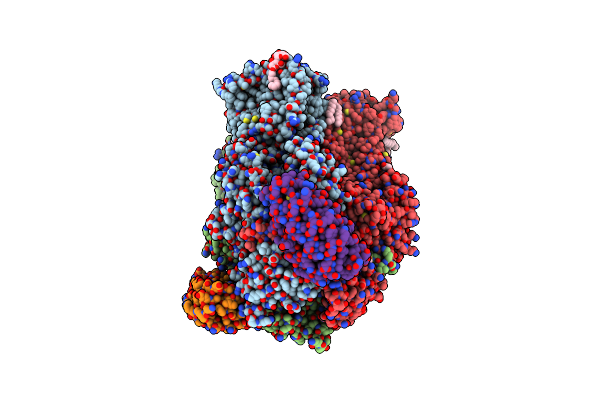
Cryo-Em Reveals Partially And Fully Assembled Native Glycine Receptors,Heteromeric Pentamer
Organism: Rattus norvegicus, Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2021-09-29 Classification: MEMBRANE PROTEIN, SIGNALING PROTEIN Ligands: HP6, DD9, LNK, MYS, D10, NBU, GLY, D12, OCT, HEX, UND |
 |
 |
Organism: Escherichia coli k-12, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2021-05-19 Classification: TRANSPORT PROTEIN Ligands: LMT, D10, EDO, DDR, MYS, 8K6, MIY, GOL, OCT, SO4, D12, PTY, C14, DDQ |
 |
Photosynthetic Reaction Center From Rhodobacter Sphaeroides Strain Rv In Surfo Crystallization
Organism: Rhodobacter sphaeroides
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2020-12-02 Classification: PHOTOSYNTHESIS Ligands: LDA, D12, MYS, PO4, EDO, NA, K, BCL, BPH, U10, D10, DIO, HTO, FE, SPN, CDL |
 |
Organism: Homo sapiens, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-11-25 Classification: MEMBRANE PROTEIN Ligands: ZMA, NA, CLR, D12, MYS, HEX, 8K6, D10, OCT, UND, ER0, TRD |
 |
Organism: Homo sapiens, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-11-25 Classification: MEMBRANE PROTEIN Ligands: ZMA, NA, CLR, D12, MYS, HEX, 8K6, D10, OCT, UND, ER0, TRD |
 |
Organism: Homo sapiens, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-11-25 Classification: MEMBRANE PROTEIN Ligands: ZMA, NA, CLR, D12, MYS, HEX, 8K6, D10, OCT, UND, ER0, TRD |
 |
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2020-08-26 Classification: MEMBRANE PROTEIN Ligands: Y01, PC1, MYS |
 |
Organism: Halobacterium salinarum (strain atcc 700922 / jcm 11081 / nrc-1)
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2019-04-24 Classification: MEMBRANE PROTEIN Ligands: RET, L2P, TRD, D10, HP6, OCT, MYS, UND, DD9, C14 |
 |
Organism: Halobacterium salinarum (strain atcc 700922 / jcm 11081 / nrc-1)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2019-04-24 Classification: MEMBRANE PROTEIN Ligands: RET, L2P, TRD, D10, HP6, OCT, MYS, UND, DD9, C14 |
 |
Organism: Halobacterium salinarum (strain atcc 700922 / jcm 11081 / nrc-1)
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2019-04-24 Classification: MEMBRANE PROTEIN Ligands: RET, L2P, TRD, D10, HP6, OCT, MYS, UND, DD9, C14 |
 |
Bacteriorhodopsin, 1 Ps State, Real-Space Refined Against 15% Extrapolated Map
Organism: Halobacterium salinarum (strain atcc 700922 / jcm 11081 / nrc-1)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2019-04-24 Classification: MEMBRANE PROTEIN Ligands: RET, L2P, TRD, D10, HP6, OCT, MYS, UND, DD9, C14 |
 |
Bacteriorhodopsin, 3 Ps State, Real-Space Refinemed Against 10% Extrapolated Map
Organism: Halobacterium salinarum (strain atcc 700922 / jcm 11081 / nrc-1)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2019-04-24 Classification: MEMBRANE PROTEIN Ligands: RET, L2P, TRD, D10, HP6, OCT, MYS, UND, DD9, C14 |
 |
Bacteriorhodopsin, 10 Ps State, Real-Space Refined Against 10% Extrapolated Map
Organism: Halobacterium salinarum (strain atcc 700922 / jcm 11081 / nrc-1)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2019-04-24 Classification: MEMBRANE PROTEIN Ligands: RET, L2P, TRD, D10, HP6, OCT, MYS, UND, DD9, C14 |
 |
Bacteriorhodopsin, 240Fs State, Real-Space Refined Against 10% Extrapolated Map
Organism: Halobacterium salinarum (strain atcc 700922 / jcm 11081 / nrc-1)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2019-04-24 Classification: MEMBRANE PROTEIN Ligands: RET, L2P, TRD, D10, HP6, OCT, MYS, UND, DD9, C14 |
 |
Bacteriorhodopsin, 330 Fs State, Real-Space Refined Against 15% Extrapolated Structure Factors
Organism: Halobacterium salinarum (strain atcc 700922 / jcm 11081 / nrc-1)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2019-04-24 Classification: MEMBRANE PROTEIN Ligands: RET, L2P, TRD, D10, HP6, OCT, MYS, UND, DD9, C14 |

