Search Count: 25
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-04-24 Classification: HYDROLASE Ligands: VY8, PG0, MXE, TOE, 7PG |
 |
The Crystal Structure Of Escherichia Coli Murr In Complex With N-Acetylglucosamine-6-Phosphate
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2022-04-20 Classification: GENE REGULATION Ligands: 4QY, GOL, MXE |
 |
Organism: Janthinobacterium sp. (strain j3)
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2021-04-07 Classification: OXIDOREDUCTASE Ligands: FES, FE2, MG, PEG, PGE, PG4, P6G, 1PE, CO3, MXE, PG0 |
 |
Crystal Structure Of Urate Oxidase From Bacillus Sp. Tb-90 In The Absence From Chloride Anion At 1.44 A Resolution
Organism: Bacillus sp. (strain tb-90)
Method: X-RAY DIFFRACTION Resolution:1.44 Å Release Date: 2020-11-25 Classification: OXIDOREDUCTASE Ligands: AZA, OXY, NA, MXE, SO4 |
 |
Crystal Structure Of Urate Oxidase From Bacillus Sp. Tb-90 In The Absence From Chloride Anion At 1.44 A Resolution
Organism: Bacillus sp. (strain tb-90)
Method: X-RAY DIFFRACTION Resolution:1.46 Å Release Date: 2020-11-25 Classification: OXIDOREDUCTASE Ligands: AZA, OXY, NA, MXE, SO4 |
 |
Crystal Structure Of Urate Oxidase From Bacillus Sp. Tb-90 In The Absence From Chloride Anion At 1.62 A Resolution
Organism: Bacillus sp. (strain tb-90)
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2020-11-25 Classification: OXIDOREDUCTASE Ligands: AZA, OXY, MXE, SO4, NA |
 |
X-Ray Structure Of Danio Rerio Histone Deacetylase 6 (Hdac6) Cd2 In Complex With A Inhibitor Ss555
Organism: Danio rerio
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2020-11-04 Classification: HYDROLASE Ligands: ZN, K, N28, EDO, TOE, GOL, CL, MXE, DMS |
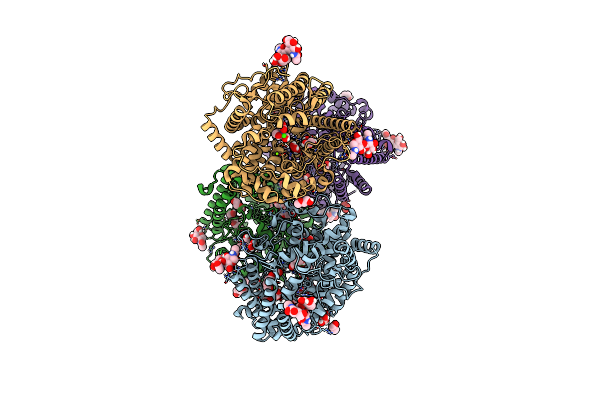 |
Crystal Structure Of The Open Conformation Of Angiotensin-1 Converting Enzyme N-Domain.
|
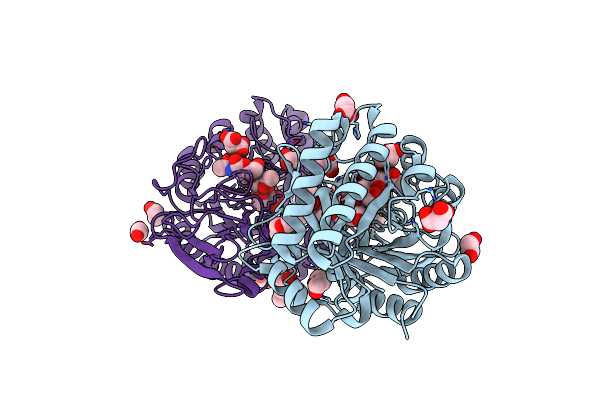 |
Crystal Structure Of The Catalytic Domain Of Chitiniphilus Shinanonensis Chitinase Chil (Cschil) Complexed With N,N'-Diacetylchitobiose
Organism: Chitiniphilus shinanonensis
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2020-09-16 Classification: HYDROLASE Ligands: ACT, MXE, MLI, PGE, EDO, NAG, PG4 |
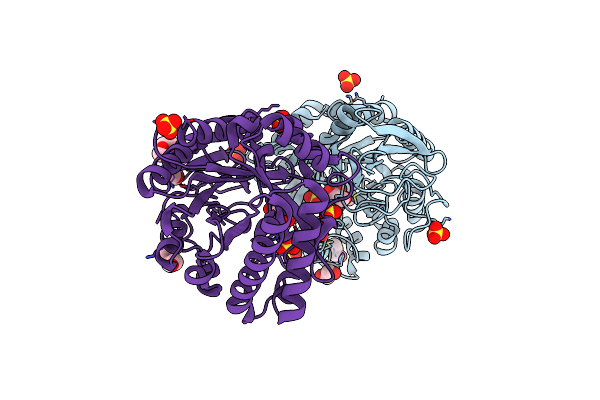 |
Crystal Structure Of The Catalytic Domain Of Chitinase Chil From Chitiniphilus Shinanonensis (Cschil)
Organism: Chitiniphilus shinanonensis
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2020-08-26 Classification: HYDROLASE Ligands: GOL, SO4, EDO, MXE, CL |
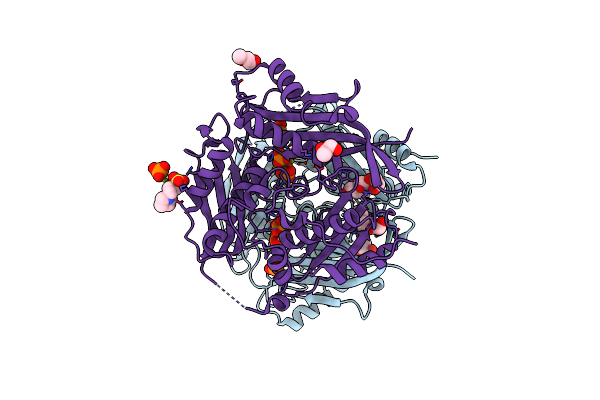 |
Crystal Structure Of Arabidopsis Thaliana S-Adenosylmethionine Synthase 2 (Atmat2)
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.52 Å Release Date: 2020-02-26 Classification: TRANSFERASE Ligands: PO4, MG, PGR, MPO, MXE, PGE |
 |
Organism: Bacillus sp. (strain tb-90)
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2018-10-10 Classification: OXIDOREDUCTASE Ligands: AZA, OXY, MXE, SO4 |
 |
Organism: Bacillus sp. (strain tb-90)
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2018-10-10 Classification: OXIDOREDUCTASE Ligands: AZA, OXY, SO4, K, MXE |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2017-01-25 Classification: DE NOVO PROTEIN Ligands: MXE, CL, PG4, PG6, NA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2016-09-14 Classification: TRANSFERASE Ligands: VX6, MXE, MES |
 |
Organism: Sordaria macrospora (strain atcc mya-333 / dsm 997 / k(l3346) / k-hell), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2016-01-13 Classification: Hydrolase/DNA Ligands: MG, MXE, PG0 |
 |
Organism: Sordaria macrospora (strain atcc mya-333 / dsm 997 / k(l3346) / k-hell), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2016-01-13 Classification: Hydrolase/DNA Ligands: MG, MXE, PG0 |
 |
Crystal Structure Of Anthrax Protective Antigen (Membrane Insertion Loop Deleted) To 1.45-A Resolution
Organism: Bacillus anthracis
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2011-10-26 Classification: protein transport, toxin Ligands: CA, MXE |
 |
Crystal Structure Of Anthrax Protective Antigen Mutant S337C N664C And Dithiolacetone Modified To 1.8-A Resolution
Organism: Bacillus anthracis
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2011-10-26 Classification: PROTEIN TRANSPORT, TOXIN Ligands: CA, MXE, ACN |