Search Count: 107
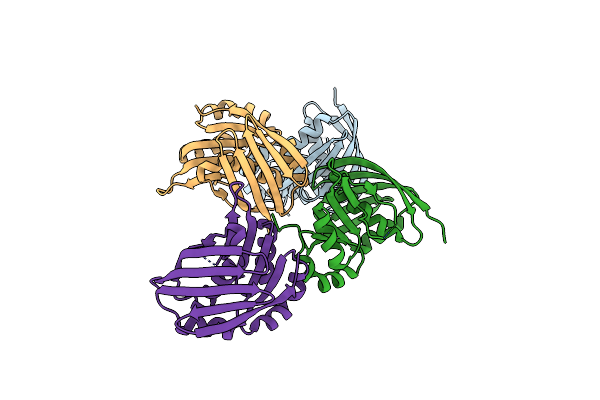 |
Crystal Structure Of The Yaeq Family Protein Vpa0551 From Vibrio Parahaemolyticus
Organism: Vibrio parahaemolyticus rimd 2210633
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2022-01-05 Classification: UNKNOWN FUNCTION |
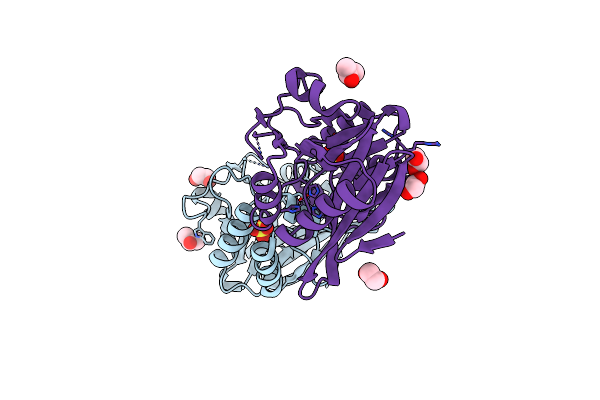 |
Crystal Structure Of The Ndm_Fim-1 Like Metallo-Beta-Lactamase From Erythrobacter Litoralis In The Mono-Zinc Form
Organism: Erythrobacter litoralis (strain htcc2594)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-01-15 Classification: HYDROLASE Ligands: ZN, SO4, IPA, EDO |
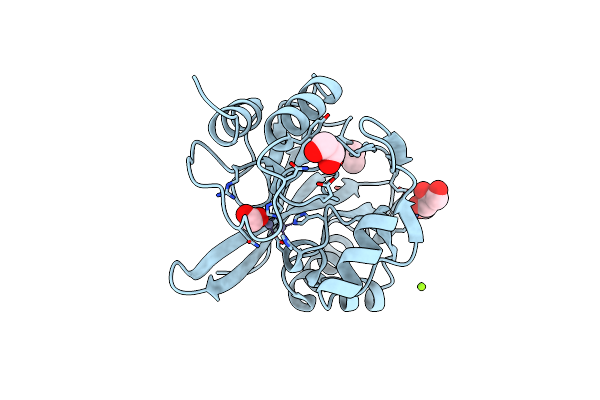 |
Organism: Hirschia baltica (strain atcc 49814 / dsm 5838 / ifam 1418)
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2019-12-25 Classification: HYDROLASE Ligands: MG, FMT, EDO, CL, ZN, GOL |
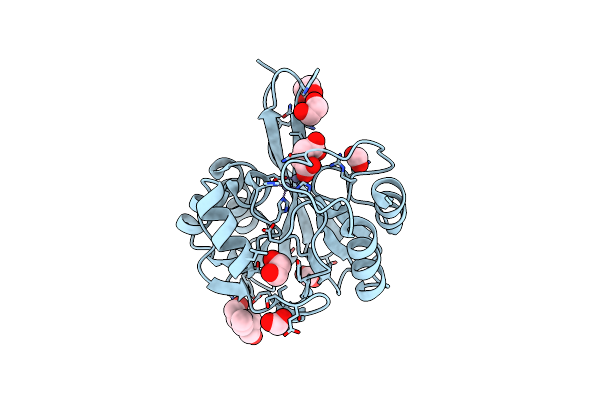 |
Crystal Structure Of Metallo Beta Lactamase From Hirschia Baltica In Complex With Succinate
Organism: Hirschia baltica (strain atcc 49814 / dsm 5838 / ifam 1418)
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2019-12-25 Classification: HYDROLASE Ligands: ZN, SIN, EDO, FMT, BTB |
 |
Crystal Structure Of Metallo Beta Lactamase From Hirschia Baltica In The Complex With The Inhibitor Captopril
Organism: Hirschia baltica (strain atcc 49814 / dsm 5838 / ifam 1418)
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2019-12-25 Classification: HYDROLASE Ligands: ZN, EDO, X8Z, NA, FMT, CL |
 |
Crystal Structure Of Metallo Beta Lactamase From Hirschia Baltica With Cadmium In The Active Site
Organism: Hirschia baltica (strain atcc 49814 / dsm 5838 / ifam 1418)
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2019-12-25 Classification: HYDROLASE Ligands: CL, EDO, CD, FMT |
 |
Crystal Structure Of Metallo Beta Lactamase From Hirschia Baltica With Nitrate In The Active Site
Organism: Hirschia baltica (strain atcc 49814 / dsm 5838 / ifam 1418)
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2019-12-25 Classification: HYDROLASE Ligands: ZN, EDO, NO3, FMT, PO4 |
 |
Organism: Erythrobacter litoralis (strain htcc2594)
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2019-12-25 Classification: HYDROLASE Ligands: CA, CL, FMT, EDO |
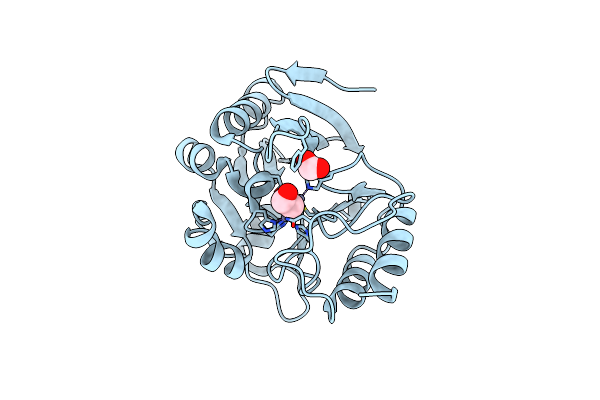 |
Crystal Structure Of Metallo Beta Lactamase From Erythrobacter Litoralis With Beta Mercaptoethanol In The Active Site
Organism: Erythrobacter litoralis (strain htcc2594)
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2019-12-25 Classification: HYDROLASE Ligands: ZN, CL, BME, FMT |
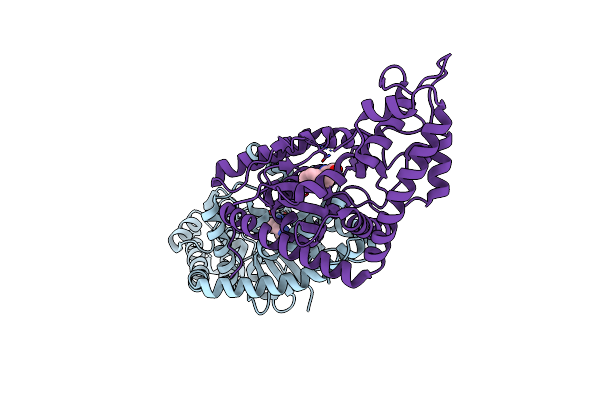 |
Organism: Haemophilus influenzae (strain atcc 51907 / dsm 11121 / kw20 / rd)
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2018-05-23 Classification: LIGASE Ligands: TRP |
 |
High Resolution Structure Of Thioredoxin-Disulfide Reductase From Vibrio Vulnificus Cmcp6 In Complex With Nadp And Fad
Organism: Vibrio vulnificus cmcp6
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2017-05-31 Classification: OXIDOREDUCTASE Ligands: NAP, FAD, K, CAC, GOL |
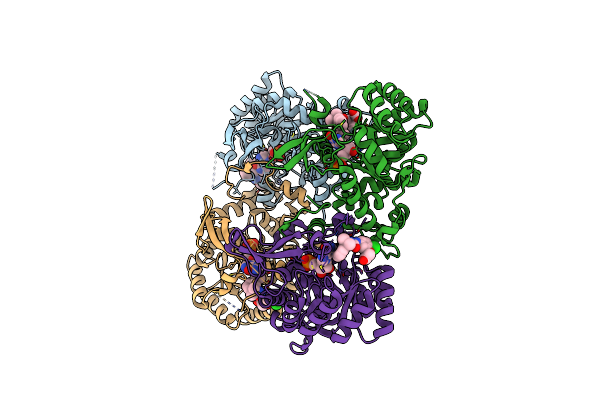 |
Crystal Structure Of Inosine 5'-Monophosphate Dehydrogenase From Clostridium Perfringens Complexed With Imp And P225
Organism: Clostridium perfringens
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2017-05-24 Classification: oxidoreductase/oxidoreductase inhibitor Ligands: IMP, 8KY |
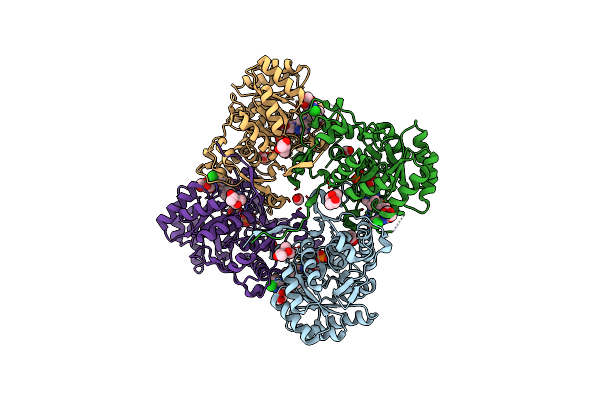 |
Crystal Structure Of Inosine 5'-Monophosphate Dehydrogenase From Clostridium Perfringens Complexed With Imp And P221
Organism: Clostridium perfringens (strain atcc 13124 / dsm 756 / jcm 1290 / ncimb 6125 / nctc 8237 / type a)
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2017-03-22 Classification: oxidoreductase/oxidoreductase inhibitor Ligands: IMP, 8N1, MPD, ACY, FMT, MRD |
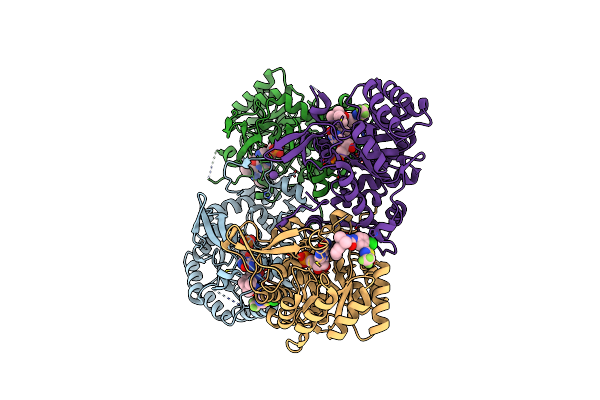 |
Crystal Structure Of Inosine 5'-Monophosphate Dehydrogenase From Clostridium Perfringens Complexed With Imp And P182
Organism: Clostridium perfringens (strain atcc 13124 / dsm 756 / jcm 1290 / ncimb 6125 / nctc 8237 / type a)
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2017-03-22 Classification: oxidoreductase/oxidoreductase inhibitor Ligands: IMP, 8L1, K, GOL |
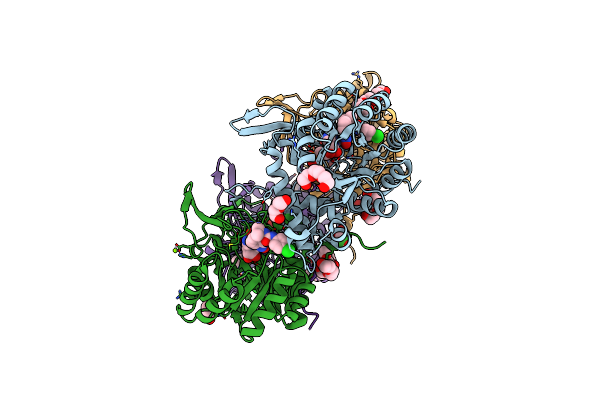 |
Crystal Structure Of Inosine 5'-Monophosphate Dehydrogenase From Clostridium Perfringens Complexed With Imp And P200
Organism: Clostridium perfringens (strain atcc 13124 / dsm 756 / jcm 1290 / ncimb 6125 / nctc 8237 / type a)
Method: X-RAY DIFFRACTION Resolution:2.37 Å Release Date: 2017-03-22 Classification: oxidoreductase/oxidoreductase inhibitor Ligands: IMP, 8L4, PGE, PEG, EDO, MG, K, PG4 |
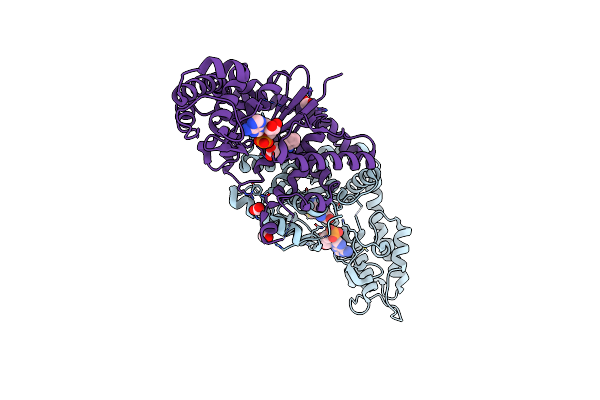 |
Crystal Structure Of Tryptophanyl-Trna Synthetase From Escherichia Coli Complexed With Amp And Tryptophan
Organism: Escherichia coli o157:h7 str. edl933
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2017-03-22 Classification: LIGASE Ligands: AMP, TRP, FMT |
 |
Crystal Structure Of The Catalytic Domain Of The Inosine Monophosphate Dehydrogenase From Bacillus Anthracis In The Complex With Imp And The Inhibitor P200
Organism: Bacillus anthracis
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2017-03-08 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: IMP, 8L4, K |
 |
Crystal Structure Of Inosine 5'-Monophosphate Dehydrogenase From Clostridium Perfringens Complexed With Imp And P178
Organism: Clostridium perfringens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2017-03-08 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: IMP, 8LA, K, MPD, FMT, SO4 |
 |
Crystal Structure Of The Catalytic Domain Of The Inosine Monophosphate Dehydrogenase From Bacillus Anthracis In The Complex With A Product Imp And The Inhibitor P182
Organism: Bacillus anthracis
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2017-03-01 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: IMP, 8L1, K, GOL |
 |
Crystal Structure Of Inosine 5'-Monophosphate Dehydrogenase From Clostridium Perfringens Complexed With Imp And P176
Organism: Clostridium perfringens
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2017-03-01 Classification: oxidoreductase/oxidoreductase inhibitor Ligands: 8L7, IMP, MPD, ACY, MRD |

