Search Count: 38
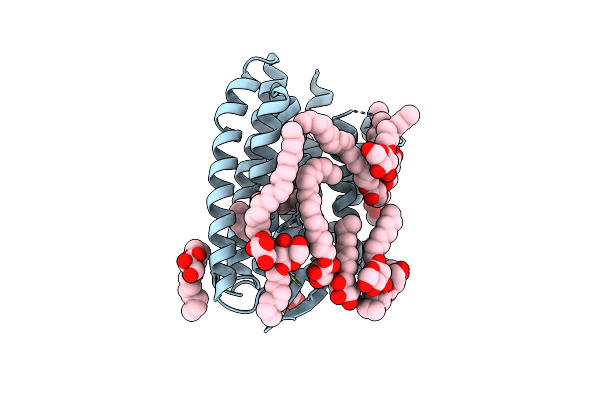 |
Grouped 150-240 Ms Dark Structure Of Sensory Rhodopsin-Ii Solved By Serial Millisecond Crystallography
Organism: Natronomonas pharaonis
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2025-04-30 Classification: SIGNALING PROTEIN Ligands: RET, CL, BOG, MPG |
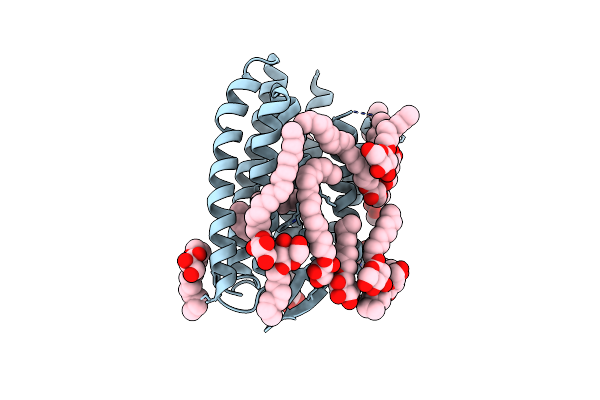 |
Continuous Dark State Structure Of Sensory Rhodopsin Ii Solved By Serial Millisecond Crystallography
Organism: Natronomonas pharaonis
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2025-04-30 Classification: SIGNALING PROTEIN Ligands: RET, CL, BOG, MPG |
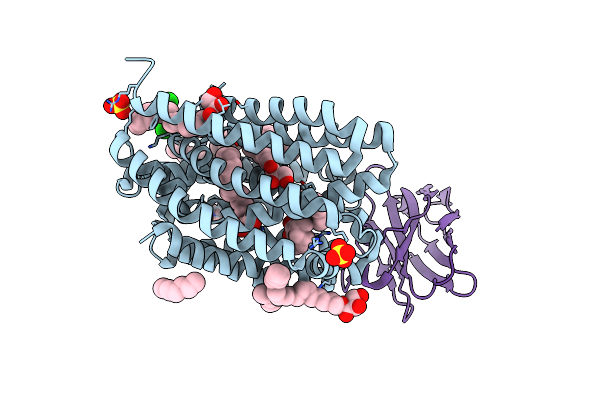 |
Lipid Iii Flippase Wzxe With Nb10 Nanobody In Outward-Facing Conformation At 2.7552 A
Organism: Escherichia coli, Lama glama
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2025-01-22 Classification: TRANSPORT PROTEIN Ligands: MPG, OCT, SO4, ZN, CL |
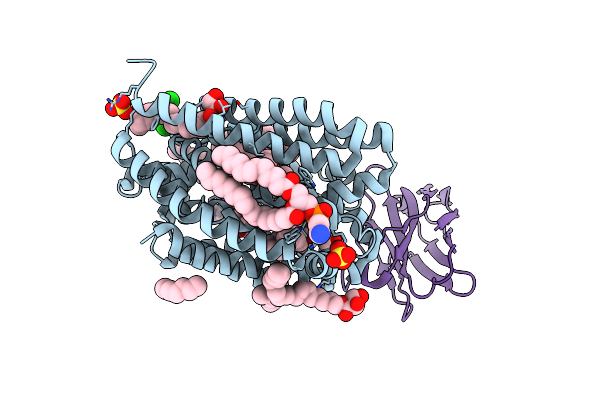 |
Lipid Iii Flippase Wzxe With Nb10 Nanobody In Outward-Facing Conformation At 0.9688 A
Organism: Escherichia coli, Lama glama
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2025-01-22 Classification: TRANSPORT PROTEIN Ligands: MPG, LOP, OCT, CL, SO4, ZN |
 |
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2025-01-15 Classification: MEMBRANE PROTEIN Ligands: MH0, MPG, OXY |
 |
Organism: Bordetella bronchiseptica rb50
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2024-11-13 Classification: MEMBRANE PROTEIN Ligands: CD, CL, MPG, HG |
 |
Organism: Bordetella bronchiseptica rb50
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2023-02-22 Classification: TRANSPORT PROTEIN Ligands: SO4, MPG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2022-06-15 Classification: MEMBRANE PROTEIN Ligands: V0S, CLR, NAG, NA, MPG |
 |
Organism: Methanocaldococcus jannaschii dsm 2661
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2021-02-03 Classification: MEMBRANE PROTEIN Ligands: LDA, PO4, MG, MPG |
 |
Organism: Methanocaldococcus jannaschii dsm 2661
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2021-02-03 Classification: MEMBRANE PROTEIN Ligands: MPG, DMA, PO4, MG, LDA |
 |
Organism: Rubrobacter xylanophilus (strain dsm 9941 / nbrc 16129)
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2020-04-08 Classification: TRANSPORT PROTEIN Ligands: MPG, RET, SO4 |
 |
Atomic Structure Of The Eukaryotic Intramembrane Ras Methyltransferase Icmt (Isoprenylcysteine Carboxyl Methyltransferase), In Complex With A Monobody
Organism: Tribolium castaneum, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2018-01-17 Classification: TRANSFERASE Ligands: SAH, UND, D10, MPG |
 |
Blastochloris Viridis Photosynthetic Reaction Center Structure Using Best Crystal Approach
Organism: Blastochloris viridis
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2017-02-08 Classification: PHOTOSYNTHESIS Ligands: HEC, DGA, BCL, BPB, MPG, FE2, MQ7, NS5, OTP, PO4 |
 |
Organism: Blastochloris viridis
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2017-02-08 Classification: PHOTOSYNTHESIS Ligands: HEC, DGA, BCL, BPB, MPG, FE2, MQ7, NS5, OTP, PO4 |
 |
Organism: Blastochloris viridis
Method: X-RAY DIFFRACTION Resolution:3.60 Å Release Date: 2017-02-08 Classification: PHOTOSYNTHESIS Ligands: HEC, DGA, BCL, BPB, MPG, FE2, MQ7, NS5, OTP, PO4 |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2016-08-17 Classification: TRANSPORT PROTEIN Ligands: VIB, MPG |
 |
Organism: Homo sapiens, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2016-07-20 Classification: MEMBRANE PROTEIN Ligands: NAG, VIS, NA, MPG |
 |
Organism: Cupriavidus metallidurans (strain atcc 43123 / dsm 2839 / nbrc 102507 / ch34)
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2016-02-17 Classification: TRANSFERASE Ligands: ZN, DSL, MPG, CL, PO4, PC, EPE |
 |
Crystal Structure Of Arnt From Cupriavidus Metallidurans Bound To Undecaprenyl Phosphate
Organism: Cupriavidus metallidurans (strain atcc 43123 / dsm 2839 / nbrc 102507 / ch34)
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2016-02-17 Classification: TRANSFERASE Ligands: MPG, EPE, PO4, CL, 5TR |
 |
Organism: Influenza a virus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2015-10-14 Classification: MEMBRANE PROTEIN Ligands: MPG |

