Search Count: 372
 |
Structure Of The Tela-Associated Type Vii Secretion System Chaperone Sir_0168
Organism: Streptococcus intermedius b196
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: CHAPERONE Ligands: EDO, CL |
 |
Organism: Streptococcus intermedius
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: CHAPERONE Ligands: FMT, ACY |
 |
Structure Of The Telb-Associated Type Vii Secretion System Chaperone Sir_0178
Organism: Streptococcus intermedius b196
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: CHAPERONE Ligands: CL |
 |
Structure Of The Tela-Associated Type Vii Secretion System Chaperone Lcpa In Complex With The Chaperone Binding Site Of Tela
Organism: Streptococcus intermedius b196
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: CHAPERONE Ligands: CIT |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: CHAPERONE Ligands: MG |
 |
Cryo-Em Structure Of The Agr2 Dimer In Complex With The Monomeric Ire1Beta Luminal Domain
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: CHAPERONE |
 |
Crystal Structure Of Enterobacter Cloacae Ycdy, A Member Of The Redox Enzyme Maturation Protein Family
Organism: Enterobacter cloacae
Method: X-RAY DIFFRACTION Release Date: 2025-06-18 Classification: CHAPERONE Ligands: CA |
 |
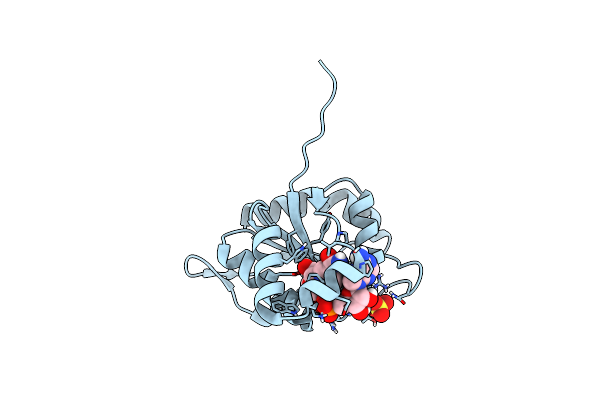
Crystal Structure Of The Nucleotide-Binding Domain Of Candida Glabrata Hsp90
Organism: Candida glabrata
Method: X-RAY DIFFRACTION Release Date: 2025-03-05 Classification: CHAPERONE |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2024-09-18 Classification: HYDROLASE Ligands: NAD, SO4 |
 |
The Structure Of Htpg M Domain In Complex With Unstructured D131D Binding Site B
Organism: Escherichia coli, Staphylococcus aureus
Method: SOLUTION NMR Release Date: 2024-06-26 Classification: CHAPERONE |
 |
The Structure Of Htpg M Domain In Complex With Unstructured D131D Binding Site A
Organism: Escherichia coli, Staphylococcus aureus
Method: SOLUTION NMR Release Date: 2024-06-26 Classification: CHAPERONE |
 |
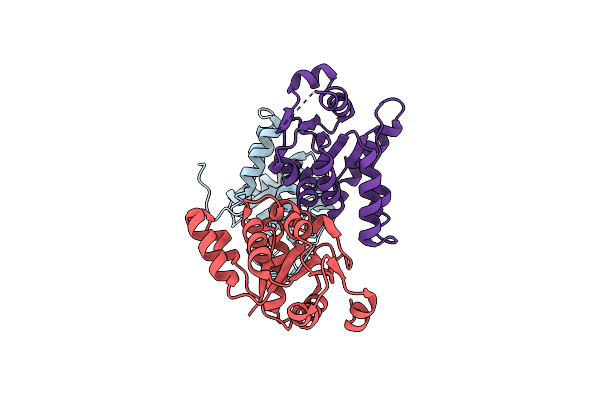
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2024-06-19 Classification: CHAPERONE Ligands: CIT |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2024-06-12 Classification: IMMUNE SYSTEM Ligands: BR |
 |
Organism: Methanocaldococcus jannaschii dsm 2661
Method: ELECTRON MICROSCOPY Release Date: 2023-12-27 Classification: CHAPERONE |
 |
Structural Basis Of Aggregate Binding/Recognition By The Aaa+ Disaggregase Clpg
Organism: Pseudomonas aeruginosa
Method: SOLUTION NMR Release Date: 2023-11-01 Classification: CHAPERONE Ligands: ZN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2023-10-11 Classification: CHAPERONE Ligands: NAG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2023-10-11 Classification: CHAPERONE Ligands: NAG |
 |
96Nm Repeat Of Human Respiratory Doublet Microtubule And Associated Axonemal Complexes
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-07-26 Classification: STRUCTURAL PROTEIN Ligands: GTP, MG, GDP |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.94 Å Release Date: 2023-07-12 Classification: CHAPERONE Ligands: W5R |
 |
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2023-07-05 Classification: CHAPERONE Ligands: ADP, NO3, MG, PO4 |

